GENOMIC
Mapping
7qE1. View the map and BAC clones (data from UCSC genome browser).
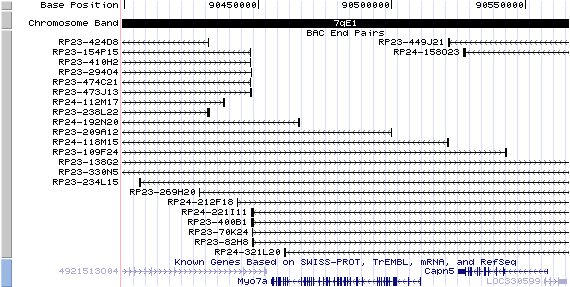
Structure
(assembly 10/03)
Isoform a (NM_008663): 47 exons, 55,781bp, Chr7: 90,454,814-90,510,594.
The figure below shows the structure of the Myo7a gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Myo7a (NM_008663), 7,135bp, view ORF and the alignment to genomic.
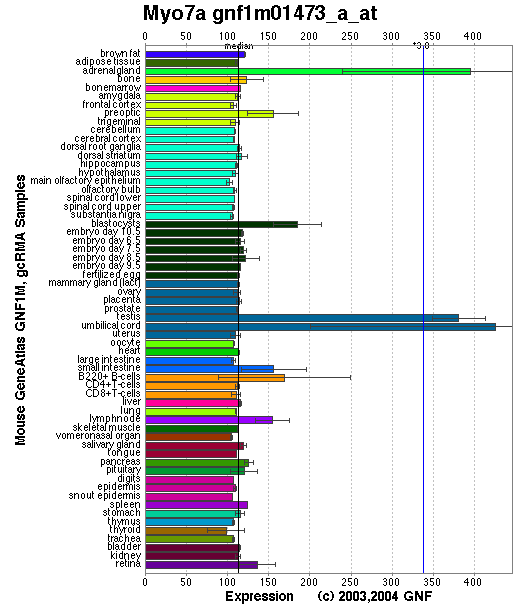
Expression Pattern
Tissue specificity: expressed in the inner ear and the retina, as well as the kidney, lung, and testis.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
Myosin VIIa (NP_032689): 2215aa, ExPaSy NiceProt view of Swiss-Prot:P97479.
Ortholog
| Species | Human | Rat | Zebrafish | Fruitfly | Worm |
| GeneView | USH1B/MYO7A | Myo5a | 252846 | CG7595/ck | hum6 |
| Protein | NP_000251 (2215aa) | NP_703203 (2177aa) | NP_694515 (2179aa) | NP_523571 (2167aa) | NP_508420 (2098aa) |
| Identities | 96% /2136aa | 96% /2133aa | 83% /1843aa | 61% /1357aa | 49% /1102aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) MYSc 59 - 742
b) IQ regions 743 - 765; 766 - 785; 812 - 834; 835 - 857
c) low complexity 865 - 900
d) low complexity 904 - 927
e) low complexity 983 - 996
g) MyTH4 1017 - 1253; 1747 - 1896
h) B41 1254 - 1469; 1898 - 2115
i) SH3 1603 - 1671
(2) Pfam domains: Pfam:MyTH4; Pfam:SH3; Pfam:RA; Pfam:IQ; Pfam:Myosin_head.
Graphic view of the Pfam domain structure.
(3) CDD domains:
a) COG4261:Talin
b) KOG4229: Myosin VII, myosin IXB and related myosins [Cell motility]
(4) Graphic view of InterPro domain structure.
(5) Transmembrane domains predicted by SOSUI: None.
Motif/Site
(1) Predicted results by ScanProsite:
a) IQ motif profile :
767 - 796: score=8.187
813 - 837: score=7.657
839 - 865: score=8.956
b) FERM domain profile :
1258 - 1605: score=33.515
1902 - 2205: score=40.712
c) Src homology 3 (SH3) domain profile :
1603 - 1672: score=9.536
d) Arginine-rich region profile : [occurs frequently]
776 - 873: score=9.842
e) Tyrosine kinase phosphorylation site : [occurs frequently]
88 - 95: Ryr.DhliY,
244 - 252: RqapDernY,
1453 - 1461: KvkeDvvnY,
2045 - 2052: KfeeDks.Y
f) ATP/GTP-binding site motif A (P-loop) : [occurs frequently]
158 - 165: GesgaGKT
g) cAMP- and cGMP-dependent protein kinase phosphorylation site : [occurs frequently]
604 - 607: RKrS,
1103 - 1106: KKtS,
1117 - 1120: KKkS,
1602 - 1605: RKrS.
h) Amidation site : [occurs frequently]
972 - 975: rGRR,
1078 - 1081: lGKK.
i) Cell attachment sequence : [occurs frequently]
1659 - 1661: RGD
j) Tyrosine sulfation site : [occurs frequently]
470 - 484:
fkleqeeYdlesidw
1712 - 1726:
ytleefsYdyfrppp
k) Bipartite nuclear targeting sequence : [occurs frequently]
1103 - 1119:
KKtsvrhklvhltlkkk
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: none
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase matched entries found, results here.
(2) ModBase predicted comparative 3D structure of P97479 from UCSC Genome Sorter.
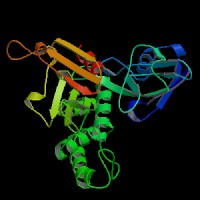
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=254,406Da, pI=8.78 (NP_000251).
FUNCTION
Ontology
a) Biological processes: visual perception; perception of sound
b) Biological process: actin filament-based movement
c) ATPase activity, coupled
d) A plus-directed microfilament motor activity
e) Component of myosin
f) Component of cytoskeleton
g) Actin binding; ATP binding; calmodulin binding.
Location
Cytoplasmic (probable). In the photoreceptor cells, mainly localized in the inner and base of outer segments as well as in the synaptic ending region.
Interaction
Myosin-VIIa may homodimerize in a two headed molecule through the formation of a coiled-coil rod to consist of two heavy chains, each containing a neck domain with five tandem IQ motifs that bind calmodulins. Myosin VIIa also binds F-actin and shows Mg2+-ATPase activity.
The Rab27a recruits distinct myosins (myosin-VIIa or myosin-Va) via different effectors (MyRIP or melanophilin) to regulate the dynamics of the melanosomes in different cell types (retinal pigment epithelium (RPE) or melanocytes). The melanophilin homolog, MyRIP, mediates the formation of a tripartite complex with RAB27A and myosin VIIa in RPE, to regulate the melanosome distribution (El-Amraoui, et al; Futter, et al). (Desnos, et al). However, the RAB27A/MyRIP protein complex does not appear to require recruitment of a myosin on the secretory granules in PC12 cells for function. Myosin VIIa is apparently not expressed in pancreatic beta cells (Waselle, et al).
Harmonin, the protein encoded by Ush1c, has been shown to bind, by means of its PDZ-domains, with the products of other Usher syndrome type 1 genes, including Myo7a, Cdh23 and Sans. The complexes formed by these protein interactions are thought to be essential for maintaining the integrity of hair cell stereocilia (Johnson, et al; Weil, et al).
PCDH15 binds to MYO7A and that both proteins are expressed in an overlapping pattern in hair bundles. PCDH15 and MYO7A cooperate to regulate the development and function of the mechanically sensitive hair bundle (Senften, et al).
No entry of the MYO7A drosophila homolog CG7595/ck shown in CuraGen interaction database.
Pathway
In retina, myosin VIIa might play a role in trafficking of ribbon-synaptic vesicle complexes and renewal of the outer photoreceptors disks. Myosin VIIa is involved in the transport of melanosomes along RPE apical processes, similarly to myosin Va in skin melanocytes (Liu, et al ). Myo7a is required for the normal processing of ingested disk membranes in the RPE, primarily in the basal transport of phagosomes into the cell body where they then fuse with lysosomes (Gibbs, et al).
In inner ear, it might maintain the rigidity of stereocilia during the dynamic movements of the bundle. Myosin VIIA participates in anchoring and holding membrane-bound elements to the actin core of the stereocilium. Myosin VIIA is therefore required for the normal gating of transducer channels (Kros, et al).Involved in hair-cell vesicle trafficking of aminoglycosides, which are known to induce ototoxicity (Richardson, et al). Myosin VIIA is a novel protein kinase A-anchoring protein.
MUTATION
Allele or SNP
10 phenotypic alleles described in MGI: 104510.
SNPs deposited in dbSNP.
Distribution
(1) Three mutations including the original sh1 mutation (Myo7ash1) are described in Gibson, et al (1995).
(2) Four mutations are described in Mburu, et al (1997).
Effect
Northern blot analysis indicated that myosin-VIIa mRNA expression, size, and stability were unaffected in the seven sh1 alleles. Immunoblot analysis showed that all seven alleles expressed some full-length myosin-VIIa protein (Hasson, et al).
PHENOTYPE
The Myoza mutations cause an autosomal recessive disorder, shaker-1 (sh1) (Gibson, et al), resembling Usher syndrome type 1B (USB1B) (OMIM:276903). The original sh1 arose spontaneously in the BALB inbred strain. The strain is described in more detail in JAX Mice database (SH1/LeJ). Homozygotes for spontaneous and chemically induced mutations exhibit deafness, hyperactivity, head-shaking, and circling, with degeneration of the organ of Corti, spiral ganglion, stria vascularis in the cochlea, and vestibular ganglion in the labyrinth (More details in Mouse Locus Catalog #Myo7a).
Mice with shaker-1 demonstrate typical neuroepithelial defects manifested by hearing loss and vestibular dysfunction but no retinal pathology. The phagocytosis of photoreceptor outer segment disks by the retinal pigment epithelium (RPE) is abnormal in Myo7a null mice. The transport of ingested disks out of the apical region is inhibited in the absence of Myo7a. The inhibited transport seems to delay phagosome-lysosomal fusion, as the degradation of ingested disks was slower in mutant RPE (Gibbs, et al).
In the Myo7a-816SB and Myo7a-6J mutants, stereocilia grow and form rows of graded heights as normal, but the bundles become progressively more disorganised. Most of these mutants show no gross electrophysiological responses, but some did show evidence of hair cell depolarisation despite the disorganisation of their bundles. In contrast, the original shaker-1 mutants, Myo7a-sh1, had normal early development of stereocilia bundles, but still showed abnormal cochlear responses (Self, et al).
Photoreceptor cell degeneration was not detected in any of the shaker1 alleles. It is argued that shaker1 mice are a useful model for testing USH1B gene therapy to retinal degeneration (Lillo, et al).
The deaf circler (dfcr) mutants are identified as mutations of the Ush1c gene, the mouse ortholog of the gene responsible for human Usher syndrome type 1C and for the non-syndromic deafness disorder DFNB18 (Johnson, et al). The waltzer mouse mutant Cdh23 phenotype was found to be similar to the retinal phenotype of Myo7a mutant mice. Mutations in the cadherin 23 gene (CDH23) cause Usher syndrome type 1D in humans (Libby, et al). The Sans gene is defective in the Jackson shaker deaf mutant, the syntenic region of Usher syndrome type 1G (Weil, et al). The products of these four genes (Ush1b/Myo7a, Ush1c/Cdh23, Ush1d/Dfnb18, and Ush1g/Sans) are thought to be in the same protein complex and therefore exhibiting similar phenotypes (Johnson, et al; Weil, et al). Mariner sensory hair cells display morphological and functional defects that are similar to those present in mouse shaker-1 hair cells, which establishes that mariner is a zebrafish model for human hereditary deafness (Ernest, et al).
By ENU mutagenesis in laboratory rat, a recessive mutant, named tornado, showing aberrant circling behavior, hyperactivity, and stereotypic head shaking. A point mutation that introduces a premature stopcodon in Myo7a was identified, revealing this mutant is an appropriate rat model for Usher syndrome type 1B (Smits, et al).
REFERENCE
- Desnos C, Schonn JS, Huet S, Tran VS, El-Amraoui A, Raposo G, Fanget I, Chapuis C, Menasche G, de Saint Basile G, Petit C, Cribier S, Henry JP, Darchen F. Rab27A and its effector MyRIP link secretory granules to F-actin and control their motion towards release sites. J Cell Biol 2003; 163: 559-70. PMID: 14610058
- El-Amraoui A, Schonn JS, Kussel-Andermann P, Blanchard S, Desnos C, Henry JP, Wolfrum U, Darchen F, Petit C. MyRIP, a novel Rab effector, enables myosin VIIa recruitment to retinal melanosomes. EMBO Rep 2002; 3: 463-70. PMID: 11964381
- Ernest S, Rauch GJ, Haffter P, Geisler R, Petit C, Nicolson T. Mariner is defective in myosin VIIA: a zebrafish model for human hereditary deafness. Hum Mol Genet 2000; 9: 2189-96. PMID: 10958658
- Futter CE, Ramalho JS, Jaissle GB, Seeliger MW, Seabra MC. The role of Rab27a in the regulation of melanosome distribution within retinal pigment epithelial cells. Mol Biol Cell 2004; 15: 2264-75. PMID: 14978221
- Gibbs D, Kitamoto J, Williams DS. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc Natl Acad Sci U S A 2003 ; 100: 6481-6. PMID: 12743369
- Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature 1995; 374: 62-4. PMID: 7870172
- Hasson T, Walsh J, Cable J, Mooseker MS, Brown SD, Steel KP. Effects of shaker-1 mutations on myosin-VIIa protein and mRNA expression. Cell Motil Cytoskeleton 1997; 37: 127-38. PMID: 9186010
- Johnson KR, Gagnon LH, Webb LS, Peters LL, Hawes NL, Chang B, Zheng QY. Mouse models of USH1C and DFNB18: phenotypic and molecular analyses of two new spontaneous mutations of the Ush1c gene. Hum Mol Genet 2003; 12: 3075-86. PMID: 14519688
- Kros CJ, Marcotti W, van Netten SM, Self TJ, Libby RT, Brown SD, Richardson GP, Steel KP. Reduced climbing and increased slipping adaptation in cochlear hair cells of mice with Myo7a mutations. Nat Neurosci 2002; 5: 41-7. PMID: 11753415
- Libby RT, Kitamoto J, Holme RH, Williams DS, Steel KP. Cdh23 mutations in the mouse are associated with retinal dysfunction but not retinal degeneration. Exp Eye Res 2003; 77: 731-9. PMID: 14609561
- Lillo C, Kitamoto J, Liu X, Quint E, Steel KP, Williams DS. Mouse models for Usher syndrome 1B. Adv Exp Med Biol 2003; 533: 143-50. PMID: 15180258
- Liu X, Ondek B, Williams DS. Mutant myosin VIIa causes defective melanosome distribution in the RPE of shaker-1 mice. Nat Genet 1998; 19: 117-8. (No Abstract)
- Mburu P, Liu XZ, Walsh J, Saw D Jr, Cope MJ, Gibson F, Kendrick-Jones J, Steel KP, Brown SD. Mutation analysis of the mouse myosin VIIA deafness gene. Genes Funct 1997; 1: 191-203. PMID: 9680294
- Richardson GP, Forge A, Kros CJ, Fleming J, Brown SD, Steel KP. Myosin VIIA is required for aminoglycoside accumulation in cochlear hair cells. J Neurosci 1997; 17: 9506-19. PMID: 9391006
- Self T, Mahony M, Fleming J, Walsh J, Brown SD, Steel KP. Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development 1998; 125: 557-66. PMID: 9435277
- Senften M, Schwander M, Kazmierczak P, Lillo C, Shin JB, Hasson T, Geleoc GS, Gillespie PG, Williams D, Holt JR, Muller U. Physical and functional interaction between protocadherin 15 and myosin VIIa in mechanosensory hair cells. J Neurosci 2006; 26: 2060-71. PMID: 16481439
- Smits BM, Peters TA, Mul JD, Croes HJ, Fransen JA, Beynon AJ, Guryev V, Plasterk RH, Cuppen E. Identification of a rat model for Usher syndrome type 1B by ENU mutagenesis-driven forward genetics. Genetics. 2005; 170: 1887-96.PMID: 15965244
- Waselle L, Coppola T, Fukuda M, Iezzi M, El-Amraoui A, Petit C, Regazzi R. Involvement of the Rab27 binding protein Slac2c/MyRIP in insulin exocytosis. Mol Biol Cell 2003; 14: 4103-13. PMID: 14517322
- Weil D, El-Amraoui A, Masmoudi S, Mustapha M, Kikkawa Y, Laine S, Delmaghani S, Adato A, Nadifi S, Zina ZB, Hamel C, Gal A, Ayadi H, Yonekawa H, Petit C. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet 2003; 12: 463-71. PMID: 12588794
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne: 07/24/2004
Updated by Wei Li: 06/25/2005
Updated by Wei Li: 04/06/2006