GENOMIC
Mapping
19qC3. View the map and BAC contig (data from UCSC genome browser).
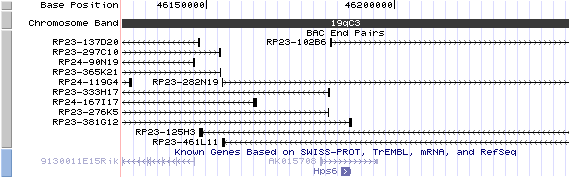
Structure
(assembly 10/03)
Hps6/NM_024747: 1 exon, 2,647 bp, chr19:46,185,636-46,188,281.
The figure shows the structure of the known isoform (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Hps6/NM_176785, 2,647bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: widely expressed.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.

PROTEIN
Sequence
Hps6p/Ru ( NP_789742): 805aa, ExPaSy NiceProt view of Swiss-Prot:Q8BLY7.
Synonym: Ruby-eye protein; Hermansky-Pudlak syndrome 6 protein homolog.
Ortholog
| Species | Mouse | Rat | Fugu | Zebrafish |
| GeneView | HPS6 | NM_181432 | BK000659 | 13581 |
| Protein | NP_789742 (805aa) | NP_852097 (809aa) | DAA00972 (916aa) | 26509 (680aa) |
| Identities | 82%/635aa | 93%/755aa | 31%/293aa | 33%/236aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) low complexity: 279-292
b) low complexity: 372-383
c) low complexity: 622-628
d) low complexity: 763-775
(2) Transmembrane domains predicted by SOSUI: none.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site [pattern] [Warning: pattern with a high probability of occurrence]:
15 - 18 NFTG .
b) Amidation site [pattern] [Warning: pattern with a high probability of occurrence]:
615 - 618 eGKR.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: XXRR-like motif in the N-terminus: KRAG
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase: none.
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.
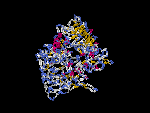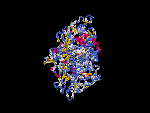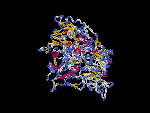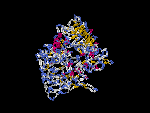
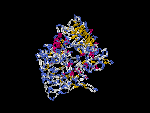
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=87,173Da, pI=6.20
FUNCTION
Ontology
a) Biological process: organelle organization and biogenesis.
b) May regulate the synthesis and function of lysosomes and of highly specialized organelles, such as melanosomes and platelet dense granules (view diagram of BLOC-2 pathway here).
c) Involved in mast cell granule secretion.
Location
Cytoplasmic.
Interaction
Zhang, et al reported that Hps6 directly interacts with Hps5 in a complex known as biogenesis of lysosome-related organelles complex-2 (or BLOC2). Gautum, et al found that Hps3 resides in the same protein complex. Interaction of the Hps5 and Hps6 proteins within the BLOC-2 is abolished by the 3aa (HCP) deletion as shown in the ru allele below. The native molecular mass of BLOC-2 was estimated to be 340 +/- 64 kDa (view diagram of BLOC-2 complex here). BLOC-2 exists in a soluble pool and associates to membranes as a peripheral membrane protein (Di Pietro, et al).
Pathway
During the maturation of melanosomes, The BLOC-2 mutants ru2 and ru are blocked between stages I and II ( Nguyen, et al ). (view diagram of melanosome blockage here). BLOC-2 functions downstream of BLOC-1 in Tyrp1 trafficking to melanosomes (Setty, et al). BLOC-2, AP-3, and AP-1 coimmunoprecipitated with Rab38 and Rab32 from MNT-1 melanocytic cell extracts,suggesting that BLOC-2, AP-3, and AP-1 proteins function in concert with Rab38 and Rab32 proteins to mediate protein trafficking to lysosome-related organelles (Bultema, et al).
MUTATION
Allele or SNP
5 phenotypic alleles described in MGI:2181763.
SNPs deposited in dbSNP.
Distribution
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 1 | 192C-257Gdel | 192C-257Gdel | L65-W86del | in-frame | ru4J (SSL/LeJ) | Zhang, et al |
| Exon 1 | 324Gins ~5.3kb IAP, 319G-324Gdup | 324Gins ~5.3kb IAP, 319G-324Gdup | M108ins ~5.3kb IAP, 319G-324Gdup | gross insertion | ru6J(C3H/HeJ) | Zhang, et al |
| Exon 1 | 559C-567Cdel | 559C-567Cdel | H187-P189del | in-frame | ru (B6) | Zhang, et al |
| Exon 1 | 1736G>A | 1736G>A | W579X | nonsense | ru3J (B6) | Zhang, et al |
(Numbering of genomic and cDNA sequence is based on the start codon of RefSeq NM_176785.)
Effect
The IAP insertion causes loss of expression of the normal kidney ru6J transcript and expression of a larger (~8.5kb) transcript in brain ( Zhang, et al ). However, no bands show up on the Western blots of brain and lung in the ru6J mutant. The two in-frame mutations in ru and ru4J are predicted non-null mutation. The nonsense mutation ru3J may not cause nonsense mediated decaying. Furthermore, the Hps6 protein also exhibits destabilization (loss or less) in Hps3 or Hps5 null mutants.
PHENOTYPE
Defects in Hps6 are the cause of ruby-eye, a mouse model of Hermansky-Pudlak syndrome type 6 (HPS-6, OMIM 607522). The strain is described in more detail in JAX Mice database (B6.Cg-Hps6ru/J). Homozygotes have hypopigmented eyes and hair, impaired secretion of lysosomal enzymes by renal proximal tubules and reduced clotting due to a platelet dense granule defect (Novak, et al)(for more details of the ru phenotype, please see the Mouse Locus Catalog #Hps6). Melanosomes of the retinal pigment epithelium (RPE) of the BLOC-2 mutants Hps5/ru and Hps6/ru2 are similar, which are greatly reduced in number and those remaining appear to be degradative forms. Melanosomes of the choroid are smaller, and some are found grouped within distinctive membrane-limited organelles (Zhang, et al). The ru2 and ru mice are completely indistinguishable in appearance. In addition, Hps5/ru2-Hps6/ru and Hps3/coa-Hps6/ru doubly homozygous mice present phenotypes identical to that of the singly homozygous mutants (Gautum, et al). Another subcellular organelle, the mast cell granule, undergoes unregulated 'kiss-and-run' fusion at the plasma membrane of mast cells of ru mice.
REFERENCE
- Bultema JJ, Ambrosio AL, Burek CL, Di Pietro SM. BLOC-2, AP-3, and AP-1 proteins function in concert with Rab38 and Rab32 proteins to mediate protein trafficking to lysosome-related organelles. J Biol Chem 2012; 287: 19550-63. PMID: 22511774
- Di Pietro SM, Falcon-Perez JM, Dell'Angelica EC. Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic 2004; 5: 276-83. PMID: 15030569
- Gautam R, Chintala S, Li W, Zhang Q, Tan J, Novak EK, Di Pietro SM, Dell'Angelica EC, Swank RT. The Hermansky-Pudlak syndrome 3 (cocoa) protein is a component of the biogenesis of lysosome-related organelles complex-2 (BLOC-2). J Biol Chem 2004; 279: 12935-42. PMID: 14718540
- Nguyen T, Novak EK, Kermani M, Fluhr J, Peters LL, Swank RT, Wei ML. Melanosome morphologies in murine models of hermansky-pudlak syndrome reflect blocks in organelle development. J Invest Dermatol 2002; 119: 1156-64. PMID: 12445206
- Novak EK, Wieland F, Jahreis GP, Swank RT. Altered secretion of kidney lysosomal enzymes in the mouse pigment mutants ruby-eye, ruby-eye-2-J, and maroon. Biochem Genet 1980; 18: 549-61. PMID: 6776948
- Setty SR, Tenza D, Truschel ST, Chou E, Sviderskaya EV, Theos AC, Lamoreux ML, Di Pietro SM, Starcevic M, Bennett DC, Dell'Angelica EC, Raposo G, Marks MS. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell 2007; 18: 768-80. PMID: 17182842
- Zhang Q, Zhao B, Li W, Oiso N, Novak EK, Rusiniak ME, Gautam R, Chintala S, O'Brien EP, Zhang Y, Roe BA, Elliott RW, Eicher EM, Liang P, Kratz C, Legius E, Spritz RA, O'Sullivan TN, Copeland NG, Jenkins NA, Swank RT. Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat Genet 33: 145-54, 2003. PubMed ID : 12548288
EDIT HISTORY:
Created by Wei Li & Jonathan W. Bourne: 06/18/2004
Updated by Wei Li: 07/28/2012