GENOMIC
Mapping
2qH4. View the map and BAC clones (data from UCSC genome browser).
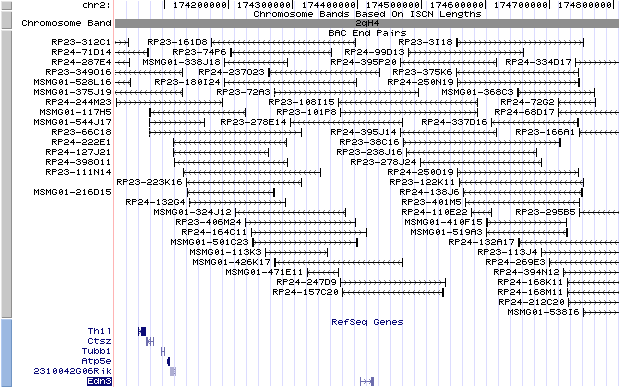
Structure
(assembly 02/2006)
Edn3(NM_007903): 5 exons, 23,270 bp, chr2:174403712-174426981.
The figure below shows the structure of the Edn3 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Edn3, seq2=human EDN3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Edn3 ( NM_007903), 2,964 bp, view ORF and the alignment to genomic.
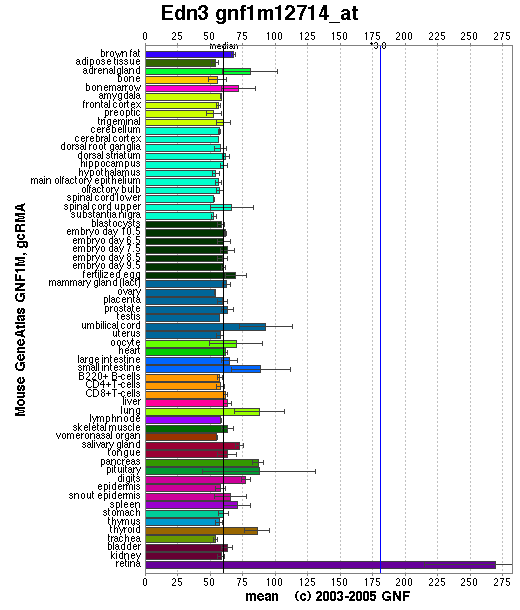
Expression Pattern
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
Browse more information in Entrez Gene, UCSC Gene Sorter, MGI.
PROTEIN
Sequence
endothelin 3 (NP_031929): 214 aa, UniProtKB/Swiss-Prot entry P48299.
Ortholog
Species
Human Chimpanzee Dog Rat GeneView
EDN3
EDN3
PPET3
Edn3
Protein
NP_996915 (219 aa)
XP_525373 (219 aa)
NP_001002942 (198 aa)
NP_001071118 (167 aa)
Identities
137/226 (60%) 137/226 (60%) 114/183 (62%) 140/214 (65%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) signal peptide: 1-16
b) END: 96-117
c) END: 158-179
d) low complexity: 185-196
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 95 to 98 RRCT.
b) Protein kinase C phosphorylation site:
Site : 28 to 30 SGR.
Site : 42 to 44 SER.
Site : 80 to 82 SGK.
Site : 101 to 103 TYK.
Site : 135 to 137 SLR.
Site : 183 to 185 SGR.
Site : 208 to 210 TDK.
c) Casein kinase II phosphorylation site:
Site : 101 to 104 TYKD.
Site : 206 to 209 SRTD.
d) Tyrosine kinase phosphorylation site:
Site : 103 to 109 KDKECVY.
Site : 103 to 110 KDKECVYY.
Site : 175 to 182 RTRDVTSY.
e) N-myristoylation site:
Site : 10 to 15 GLTVTS.
Site : 41 to 46 GSERGC.
Site : 45 to 50 GCEETV.
f) Amidation site:
Site : 137 to 140 RGKR.
g) Endothelin family signature:
Site : 97 to 111 CTCFTYKDKECVYYC.
Site : 159 to 173 CTCMGADDKACAHFC.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 1
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KAHR
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
ModBase predicted 3D structure of P48299 from UCSC Gene Sorter:
 From left to right: Front, Top, and Side views of predicted protein.
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=23,322.2Da, pI=8.54.
| Species | Human | Chimpanzee | Dog | Rat |
| GeneView | EDN3 | EDN3 | PPET3 | Edn3 |
| Protein | NP_996915 (219 aa) | XP_525373 (219 aa) | NP_001002942 (198 aa) | NP_001071118 (167 aa) |
| Identities | 137/226 (60%) | 137/226 (60%) | 114/183 (62%) | 140/214 (65%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) signal peptide: 1-16
b) END: 96-117
c) END: 158-179
d) low complexity: 185-196
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 95 to 98 RRCT.
b) Protein kinase C phosphorylation site:
Site : 28 to 30 SGR.
Site : 42 to 44 SER.
Site : 80 to 82 SGK.
Site : 101 to 103 TYK.
Site : 135 to 137 SLR.
Site : 183 to 185 SGR.
Site : 208 to 210 TDK.
c) Casein kinase II phosphorylation site:
Site : 101 to 104 TYKD.
Site : 206 to 209 SRTD.
d) Tyrosine kinase phosphorylation site:
Site : 103 to 109 KDKECVY.
Site : 103 to 110 KDKECVYY.
Site : 175 to 182 RTRDVTSY.
e) N-myristoylation site:
Site : 10 to 15 GLTVTS.
Site : 41 to 46 GSERGC.
Site : 45 to 50 GCEETV.
f) Amidation site:
Site : 137 to 140 RGKR.
g) Endothelin family signature:
Site : 97 to 111 CTCFTYKDKECVYYC.
Site : 159 to 173 CTCMGADDKACAHFC.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 1
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KAHR
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
ModBase predicted 3D structure of P48299 from UCSC Gene Sorter:


 From left to right: Front, Top, and Side views of predicted protein.
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=23,322.2Da, pI=8.54.
FUNCTION
Ontology
(1) Biological process: melanocyte differentiation, neural crest cell migration, cell-cell signaling, signal transduction, smooth muscle contraction, sensory perception of sound.
(2) Hormone secretion.
(3) Receptor binding.
(4) Inositol phosphate-mediated signaling.
(5) Regulation of vasoconstriction.
Location
Extracellular space.
Interaction
The protein encoded by this gene is a member of the endothelin family. Endothelins are endothelium-derived vasoactive peptides involved in a variety of biological functions. The active form of this protein is a 21 amino acid peptide processed from the precursor protein. The active peptide is a ligand for endothelin receptor type B (EDNRB). The interaction of this endothelin with EDNRB is essential for development of neural crest-derived cell lineages, such as melanocytes and enteric neurons.
View interactions in HPRD
View co-occured partners in literature searched by PPI Finder.
Pathway
MUTATION
Allele or SNP
3 phenotypic alleles of ls are described in MGI:95285.
SNPs deposited in dbSNP Build 128.
Distribution
The 3 phenotypic alleles include 1 targeted KO, 1 spontaneous, and 1 chemically induced.
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_007903. view ORF here.)
Effect
A ENU-derived Edn3 mutant (R96H) is predicted to disrupt furin-mediated proteolytic cleavage of pro-endothelin that is necessary to form biologically active EDN3 (Matera et al.).
PHENOTYPE
Endothelin 3 (Edn3) encodes a ligand important to developing neural crest cells and is allelic to the spontaneous mouse mutation occurring at the lethal spotting (ls) locus. Edn3(ls/ls) mutants exhibit a spotted phenotype due to reduced numbers of neural crest-derived melanocyte precursors in the skin. Homozygotes for mutations at this locus exhibit aganglionic megacolon with white spotting of the hair coat due to impaired expansion and differentiation of epidermal melanoblasts. Mutants die around weaning with impacted colons.
REFERENCE
- Matera I, Cockroft JL, Moran JL, Beier DR, Goldowitz D, Pavan WJ. A mouse model of Waardenburg syndrome type IV resulting from an ENU-induced mutation in endothelin 3. Pigment Cell Res 2007; 20:210-5. PMID: 17516928