GENOMIC
Mapping
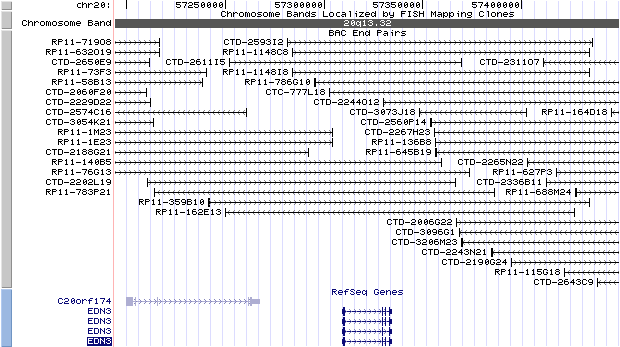
20q13.32. View the map and BAC clones (data from UCSC genome browser).
Structure
(assembly 03/2006)
isoform 1 (NM_207034): 5 exons, 25,548 bp, chr20:57308894-57334441.
isoform 2 (NM_207032): 5 exons, 25,548 bp, chr20:57308894-57334441.
isoform 3 (NM_207033): 4 exons, 25,548 bp, chr20:57308894-57334441.
Four alternatively spliced transcript variants encoding three distinct isoforms have been observed. Variant (1) encodes the longest isoform 1 (NM_000114). Variant (4), NM_207034, contains an additional fragment within the 3' UTR compared to variant (1). Both variants encode the same isoform 1. Variant (2) uses an alternative splice site in the coding region and lacks a segment in the 3' UTR compared to variant (1). The resulting isoform 2 has a distinct and shorter C-terminus, as compared to isoform 1. Variant (3) lacks an exon and uses an alternative splice site in the coding region compared to variant (1). The translation frame remains the same. The resulting isoform 3 lacks an internal region, as compared to isoform 1.
The figure below shows the structure of the EDN3 gene (data from UCSC genome browser).
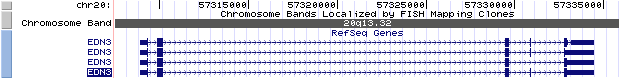
Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Edn3, seq2=human EDN3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
a) isoform 1 ( NM_207034), 2,659 bp, view ORF and the alignment to genomic.
b) isoform 2 ( NM_207032), 2,694 bp, view ORF and the alignment to genomic.
c) isoform 3 ( NM_207033), 2,617 bp, view ORF and the alignment to genomic.
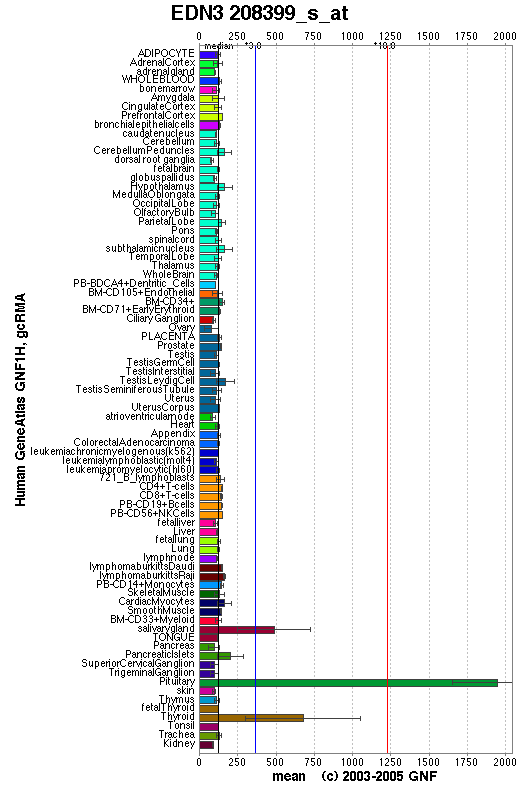
Expression Pattern
Expressed in trophoblasts and placental stem villi vessels, but not in cultured placental smooth muscle cells.
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
View more information in Entrez Gene, or UCSC Gene Sorter, or GeneCards.
PROTEIN
Sequence
a) endothelin 3 isoform 1 preproprotein (NP_996917): 238 aa, UniProtKB/Swiss-Prot entry P14138. Note: NM_000114 also endodes isoform 1 (NP_000105).
b) endothelin 3 isoform 2 preproprotein (NP_996915): 219 aa, UniProtKB/Swiss-Prot entry P14138.
c) endothelin 3 isoform 3 preproprotein (NP_996916): 224 aa, UniProtKB/Swiss-Prot entry Q4FAT2.
Ortholog (isoform 2)
Species
Chimpanzee Dog Mouse Rat GeneView
EDN3
PPET3
Edn3
Edn3
Protein
XP_525373 (219 aa)
NP_001002942 (198 aa)
NP_031929 (214 aa)
NP_001071118 (167 aa)
Identities
217/219 (99%) 137/212 (64%) 137/226 (60%) 108/215 (50%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain (isoform 2)
(1) Domains predicted by SMART:
a) signal peptide: 1-16
b) END: 96-117
c) END: 158-179
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 95 to 98 RRCT.
b) Protein kinase C phosphorylation site:
Site : 94 to 96 SRR.
Site : 101 to 103 TYK.
Site : 135 to 137 SFR.
Site : 152 to 154 SHR.
Site : 189 to 191 TDK.
Site : 213 to 215 TNK.
c) Casein kinase II phosphorylation site:
Site : 42 to 45 SEGD.
Site : 101 to 104 TYKD.
Site : 189 to 192 TDKE.
d) Tyrosine kinase phosphorylation site:
Site : 103 to 109 KDKECVY.
Site : 103 to 110 KDKECVYY.
e) N-myristoylation site:
Site : 10 to 15 GLTVTS.
Site : 80 to 85 GQEQAA.
Site : 134 to 139 GSFRGK.
Site : 199 to 204 GANRGL.
f) Amidation site:
Site : 28 to 31 AGRR.
Site : 137 to 140 RGKR.
g) Microbodies C-terminal targeting signal:
Site : 217 to 219 SRL.
h) Endothelin family signature:
Site : 97 to 111 CTCFTYKDKECVYYC.
Site : 159 to 173 CACVGRYDKACLHFC.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 1
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KASR
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of P14138 from UCSC Gene Sorter:
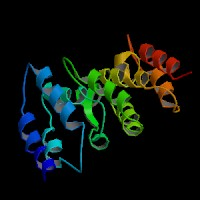
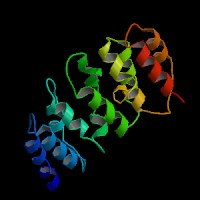
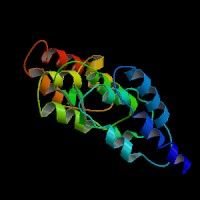
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=23,596 Da, pI=8.65 (isoform 2).
FUNCTION
Ontology
(1) Biological process: cell-cell signaling, signal transduction, smooth muscle contraction, sensory perception of sound.
(2) Hormone secretion.
(3) Receptor binding.
(4) Inositol phosphate-mediated signaling.
(5) Regulation of vasoconstriction.
Location
Extracellular space.
Interaction
The protein encoded by this gene is a member of the endothelin family. Endothelins are endothelium-derived vasoactive peptides involved in a variety of biological functions. The active form of this protein is a 21 amino acid peptide processed from the precursor protein. The active peptide is a ligand for endothelin receptor type B (EDNRB). The interaction of this endothelin with EDNRB is essential for development of neural crest-derived cell lineages, such as melanocytes and enteric neurons.
View interactions in HPRD
View co-occured partners in literature searched by PPI Finder.
Pathway
MUTATION
Allele or SNP
11 mutations deposited in HGMD.
SNPs deposited in dbSNP.
Selected allelic examples described in OMIM.
Distribution
Except the mutations deposited in HGMD, Garcia-Barcelo, et al (2004) found two novel mutations in the EDN3 gene: a missense mutation (E48D) in exon 2 resulting from a G>C transversion; and a de novo C>A transversion at nucleotide -19 from the start codon (5'UT -19C>A), which could be affecting as yet unexplored regulatory regions. E48D was found in a patient who also harbored a deletion in RET and was affected with TCA.
Note that the numbering of the mutations in HGMD is based on EDN3 cDNA (NM_000114), view ORF here.
Effect
PHENOTYPE
Defects in EDN3 are the cause of Hirschsprung disease type 1 (HSCR1) [MIM:142623]; also known as aganglionic megacolon. This genetic disorder of neural crest development is characterized by the absence of intramural ganglion cells in the hindgut; often resulting in intestinal obstruction.
Defects in EDN3 are a cause of congenital central hypoventilation syndrome (CCHS) [MIM:209880]; also known as congenital failure of autonomic control or Ondine curse. CCHS is a rare disorder characterized by abnormal control of respiration in the absence of neuromuscular or lung disease, or an identifiable brain stem lesion. A deficiency in autonomic control of respiration results in inadequate or negligible ventilatory and arousal responses to hypercapnia and hypoxemia.
Defects in EDN3 are a cause of Waardenburg syndrome type IV (WS4) [MIM:277580]; also known as Waardenburg-Shah syndrome. WS4 is characterized by the association of Waardenburg features (depigmentation and deafness) and the absence of enteric ganglia in the distal part of the intestine (Hirschsprung disease).
REFERENCE
- Garcia-Barcelo M, Sham MH, Lee WS, Lui VC, Chen BL, Wong KK, Wong JS, Tam PK.
Highly recurrent RET mutations and novel mutations in genes of the receptor tyrosine kinase and endothelin receptor B pathways in Chinese patients with sporadic Hirschsprung disease. Clin Chem 2004; 50:93-100. PMID: 14633923
| Species | Chimpanzee | Dog | Mouse | Rat |
| GeneView | EDN3 | PPET3 | Edn3 | Edn3 |
| Protein | XP_525373 (219 aa) | NP_001002942 (198 aa) | NP_031929 (214 aa) | NP_001071118 (167 aa) |
| Identities | 217/219 (99%) | 137/212 (64%) | 137/226 (60%) | 108/215 (50%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain (isoform 2)
(1) Domains predicted by SMART:
a) signal peptide: 1-16
b) END: 96-117
c) END: 158-179
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 95 to 98 RRCT.
b) Protein kinase C phosphorylation site:
Site : 94 to 96 SRR.
Site : 101 to 103 TYK.
Site : 135 to 137 SFR.
Site : 152 to 154 SHR.
Site : 189 to 191 TDK.
Site : 213 to 215 TNK.
c) Casein kinase II phosphorylation site:
Site : 42 to 45 SEGD.
Site : 101 to 104 TYKD.
Site : 189 to 192 TDKE.
d) Tyrosine kinase phosphorylation site:
Site : 103 to 109 KDKECVY.
Site : 103 to 110 KDKECVYY.
e) N-myristoylation site:
Site : 10 to 15 GLTVTS.
Site : 80 to 85 GQEQAA.
Site : 134 to 139 GSFRGK.
Site : 199 to 204 GANRGL.
f) Amidation site:
Site : 28 to 31 AGRR.
Site : 137 to 140 RGKR.
g) Microbodies C-terminal targeting signal:
Site : 217 to 219 SRL.
h) Endothelin family signature:
Site : 97 to 111 CTCFTYKDKECVYYC.
Site : 159 to 173 CACVGRYDKACLHFC.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 1
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KASR
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of P14138 from UCSC Gene Sorter:



From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=23,596 Da, pI=8.65 (isoform 2).
(1) Domains predicted by SMART:
a) signal peptide: 1-16
b) END: 96-117
c) END: 158-179
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 95 to 98 RRCT.
b) Protein kinase C phosphorylation site:
Site : 94 to 96 SRR.
Site : 101 to 103 TYK.
Site : 135 to 137 SFR.
Site : 152 to 154 SHR.
Site : 189 to 191 TDK.
Site : 213 to 215 TNK.
c) Casein kinase II phosphorylation site:
Site : 42 to 45 SEGD.
Site : 101 to 104 TYKD.
Site : 189 to 192 TDKE.
d) Tyrosine kinase phosphorylation site:
Site : 103 to 109 KDKECVY.
Site : 103 to 110 KDKECVYY.
e) N-myristoylation site:
Site : 10 to 15 GLTVTS.
Site : 80 to 85 GQEQAA.
Site : 134 to 139 GSFRGK.
Site : 199 to 204 GANRGL.
f) Amidation site:
Site : 28 to 31 AGRR.
Site : 137 to 140 RGKR.
g) Microbodies C-terminal targeting signal:
Site : 217 to 219 SRL.
h) Endothelin family signature:
Site : 97 to 111 CTCFTYKDKECVYYC.
Site : 159 to 173 CACVGRYDKACLHFC.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 1
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KASR
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of P14138 from UCSC Gene Sorter:



From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=23,596 Da, pI=8.65 (isoform 2).
FUNCTION
Ontology
(1) Biological process: cell-cell signaling, signal transduction, smooth muscle contraction, sensory perception of sound.
(2) Hormone secretion.
(3) Receptor binding.
(4) Inositol phosphate-mediated signaling.
(5) Regulation of vasoconstriction.
Location
Extracellular space.
Interaction
The protein encoded by this gene is a member of the endothelin family. Endothelins are endothelium-derived vasoactive peptides involved in a variety of biological functions. The active form of this protein is a 21 amino acid peptide processed from the precursor protein. The active peptide is a ligand for endothelin receptor type B (EDNRB). The interaction of this endothelin with EDNRB is essential for development of neural crest-derived cell lineages, such as melanocytes and enteric neurons.
View interactions in HPRD
View co-occured partners in literature searched by PPI Finder.
Pathway
MUTATION
Allele or SNP
11 mutations deposited in HGMD.
SNPs deposited in dbSNP.
Selected allelic examples described in OMIM.
Distribution
Except the mutations deposited in HGMD, Garcia-Barcelo, et al (2004) found two novel mutations in the EDN3 gene: a missense mutation (E48D) in exon 2 resulting from a G>C transversion; and a de novo C>A transversion at nucleotide -19 from the start codon (5'UT -19C>A), which could be affecting as yet unexplored regulatory regions. E48D was found in a patient who also harbored a deletion in RET and was affected with TCA.
Note that the numbering of the mutations in HGMD is based on EDN3 cDNA (NM_000114), view ORF here.
Effect
PHENOTYPE
Defects in EDN3 are the cause of Hirschsprung disease type 1 (HSCR1) [MIM:142623]; also known as aganglionic megacolon. This genetic disorder of neural crest development is characterized by the absence of intramural ganglion cells in the hindgut; often resulting in intestinal obstruction.
Defects in EDN3 are a cause of congenital central hypoventilation syndrome (CCHS) [MIM:209880]; also known as congenital failure of autonomic control or Ondine curse. CCHS is a rare disorder characterized by abnormal control of respiration in the absence of neuromuscular or lung disease, or an identifiable brain stem lesion. A deficiency in autonomic control of respiration results in inadequate or negligible ventilatory and arousal responses to hypercapnia and hypoxemia.
Defects in EDN3 are a cause of Waardenburg syndrome type IV (WS4) [MIM:277580]; also known as Waardenburg-Shah syndrome. WS4 is characterized by the association of Waardenburg features (depigmentation and deafness) and the absence of enteric ganglia in the distal part of the intestine (Hirschsprung disease).
REFERENCE
- Garcia-Barcelo M, Sham MH, Lee WS, Lui VC, Chen BL, Wong KK, Wong JS, Tam PK. Highly recurrent RET mutations and novel mutations in genes of the receptor tyrosine kinase and endothelin receptor B pathways in Chinese patients with sporadic Hirschsprung disease. Clin Chem 2004; 50:93-100. PMID: 14633923