GENOMIC
Mapping
10qD1. View the map and BAC clones (data from UCSC genome browser).
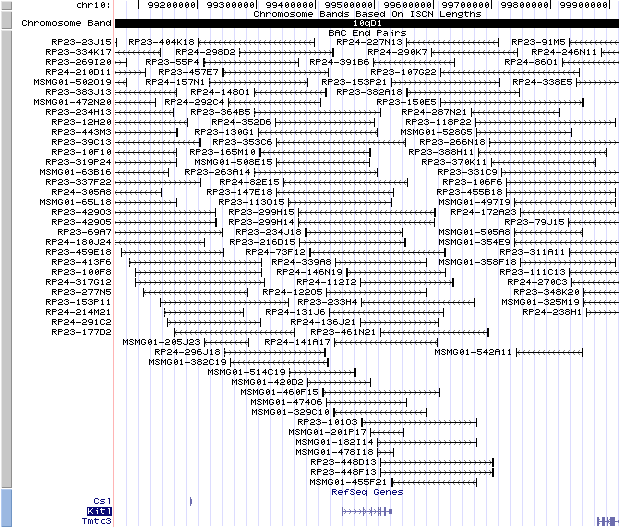
Structure
(assembly 02/2006)
Kitl (NM_013598): 10 exons, 84,588 bp, chr10:99445512-99530099
The figure below shows the structure of the Kitl gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Kitl, seq2=human KITLG) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Kitl (NM_013598), 5,449 bp, view ORF and the alignment to genomic.
Expression Pattern
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
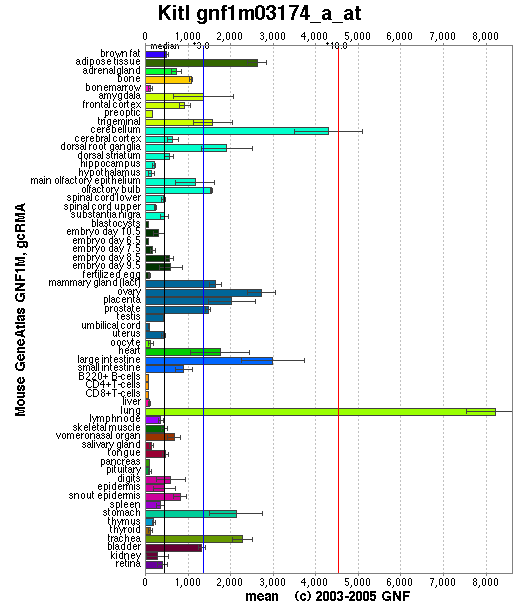
Browse more information in Entrez Gene, UCSC Gene Sorter, MGI.
PROTEIN
Sequence
kit ligand (NP_038626): 273 aa, UniProtKB/Swiss-Prot entry P20826.
Ortholog
Species
Human Chimpanzee Dog Rat Fowl GeneView
KITLG
KITLG
KITLG
Kitl
LOC396028
Protein
NP_000890 (273 aa)
XP_509255 (273 aa)
NP_001012753 (274 aa)
NP_068615 (273 aa)
NP_990461 (287 aa)
Identities
226/273 (82%) 226/273 (82%) 221/274 (80%) 261/273 (95%) 151/287 (52%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
Pfam:SCF 1-273
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a MEMBRANE PROTEIN
which have 2 transmembrane helices.
No. N terminal transmembrane region C terminal type length 1 4 TQTWIITCIYLQLLLFNPLVK 24 SECONDARY 21 2 215 WTAMALPALISLVIGFAFGAL 235 PRIMARY 21
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 90 to 93 NISE.
Site : 97 to 100 NYSI.
Site : 145 to 148 NRSI.
Site : 195 to 198 NDSS.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 239 to 242 KKQS.
c) Protein kinase C phosphorylation site:
Site : 126 to 128 SPK.
Site : 192 to 194 SLR.
Site : 200 to 202 SNR.
d) Casein kinase II phosphorylation site:
Site : 83 to 86 TLLD.
Site : 99 to 102 SIID.
Site : 136 to 139 TPEE.
e) N-myristoylation site:
Site : 30 to 35 GNPVTD.
Site : 212 to 217 GLQWTA.
(2) Predicted results of subprograms by PSORT II:
a) Seems to have no N-terminal signal peptide
b) Tentative number of TMS(s) for the threshold 0.5: 2
Number of TMS(s) for threshold 0.5: 1
INTEGRAL Likelihood = -5.89 Transmembrane 219 - 235
PERIPHERAL Likelihood = 4.61 (at 99)
ALOM score: -5.89 (number of TMSs: 1)
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
ModBase predicted 3D structure of P20826 from UCSC Gene Sorter:
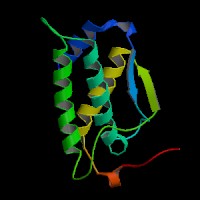
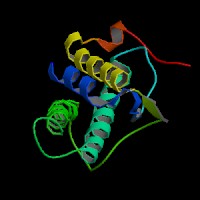
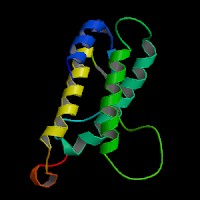
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=30,645.3Da, pI=5.28.
| Species | Human | Chimpanzee | Dog | Rat | Fowl |
| GeneView | KITLG | KITLG | KITLG | Kitl | LOC396028 |
| Protein | NP_000890 (273 aa) | XP_509255 (273 aa) | NP_001012753 (274 aa) | NP_068615 (273 aa) | NP_990461 (287 aa) |
| Identities | 226/273 (82%) | 226/273 (82%) | 221/274 (80%) | 261/273 (95%) | 151/287 (52%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
Pfam:SCF 1-273
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a MEMBRANE PROTEIN
which have 2 transmembrane helices.
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 4 | TQTWIITCIYLQLLLFNPLVK | 24 | SECONDARY | 21 |
| 2 | 215 | WTAMALPALISLVIGFAFGAL | 235 | PRIMARY | 21 |
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 90 to 93 NISE.
Site : 97 to 100 NYSI.
Site : 145 to 148 NRSI.
Site : 195 to 198 NDSS.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 239 to 242 KKQS.
c) Protein kinase C phosphorylation site:
Site : 126 to 128 SPK.
Site : 192 to 194 SLR.
Site : 200 to 202 SNR.
d) Casein kinase II phosphorylation site:
Site : 83 to 86 TLLD.
Site : 99 to 102 SIID.
Site : 136 to 139 TPEE.
e) N-myristoylation site:
Site : 30 to 35 GNPVTD.
Site : 212 to 217 GLQWTA.
(2) Predicted results of subprograms by PSORT II:
a) Seems to have no N-terminal signal peptide
b) Tentative number of TMS(s) for the threshold 0.5: 2
Number of TMS(s) for threshold 0.5: 1
INTEGRAL Likelihood = -5.89 Transmembrane 219 - 235
PERIPHERAL Likelihood = 4.61 (at 99)
ALOM score: -5.89 (number of TMSs: 1)
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
ModBase predicted 3D structure of P20826 from UCSC Gene Sorter:



2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=30,645.3Da, pI=5.28.
FUNCTION
Ontology
(1) Biological process: hemopoiesis, epithelial cell proliferation, neural crest cell migration, signal transduction, cell adhesion.
(2) Growth factor activity.
(3) Receptor binding.
(4) Regulation of pigmentation during development.
Location
Plasma membrane, cytoplasm, extracellular space.
Interaction
This gene encodes the ligand of the tyrosine-kinase receptor encoded by the KIT locus. This ligand is a pleiotropic factor that acts in utero in germ cell and neural cell development, and hematopoiesis, all believed to reflect a role in cell migration. In adults, it functions pleiotropically, while mostly noted for its continued requirement in hematopoiesis.
View interactions in HPRD
View co-occured partners in literature searched by PPI Finder.
Pathway
Stimulates the proliferation of mast cells. Able to augment the proliferation of both myeloid and lymphoid hematopoietic progenitors in bone marrow culture. Mediates also cell-cell adhesion. Acts synergistically with other cytokines, probably interleukins.
Cytokine-cytokine receptor interaction , Melanogenesis, Hematopoietic cell lineage in KEGG.
MUTATION
Allele or SNP
48 phenotypic alleles of Kitl are described in MGI:96974.
SNPs deposited in dbSNP Build 128.
Distribution
Among the reported 48 phenotypic alleles, 18 are spontaneous and 13 are chemically induced.
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_013598. view ORF here.)
Effect
Intracellular transport of Kitl to the cell surface is driven by a motif in the cytoplasmic tail that acts independently of the previously described basolateral sorting signal. Transport of Kitl to the cell surface is controlled at the level of the endoplasmic reticulum (ER) and requires a C-terminal valine residue positioned at a distance of 19-36 amino acids from the border between the transmembrane and cytoplasmic domains. Deletion or substitution of the valine with other hydrophobic amino acids results in ER accumulation and reduced cell surface transport of Kitl at physiological expression levels (Paulhe et al.).
PHENOTYPE
Mutations in this gene affect migration of embryonic stem cells and cause similar phenotypes to mutations in its receptor gene (Kit). Mutants show mild to severe defects in pigmentation, hemopoiesis and reproduction. The Sl allele arose spontaneously in the C57BL/6J strain. The strain is described in more detail in JAX Mice database (C57BL/6J-KitlSl-20J/J).
REFERENCE
- Paulhe F, Imhof BA, Wehrle-Haller B. A specific endoplasmic reticulum export signal drives transport of stem cell factor (Kitl) to the cell surface. J Biol Chem 2004; 279:55545-55. PMID: 15475566