CONREAL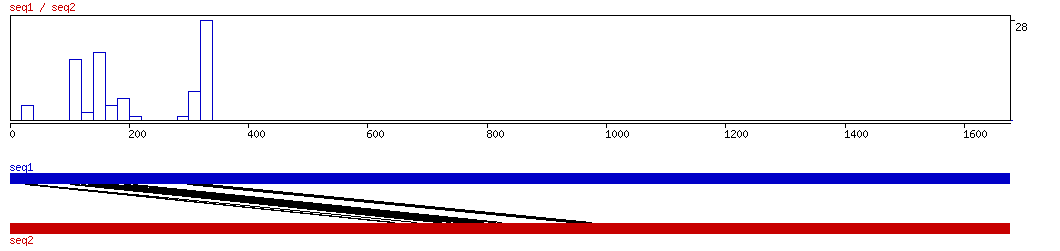
CONREAL ALIGNMENT 10 20
seq1 cgtcatggcgcgagccctcctactc---------------------------------------------------------------------------
| | | || |
seq2 ggatgagaaggaagacaatgggaggtggatgggagtatgacacacaggaaggcacaaaggaacacataagtggtggtaactattattgattacaatattt
10 20 30 40 50 60 70 80 90 100
seq1 ----------------------------------------------------------------------------------------------------
seq2 cagtgatcccagcgccaccttccacattaccgggtggggtctttactcattacaggggcaaatccatactaaaaaactcactgtgtgctggggtgtagcg
110 120 130 140 150 160 170 180 190 200
seq1 ----------------------------------------------------------------------------------------------------
seq2 cgatcagcacgcactttatccagcacacaggcagaggggacagatgagaccgggtatggttgagggtgcattttctaaggaggaaacggaaggataggaa
210 220 230 240 250 260 270 280 290 300
seq1 ----------------------------------------------------------------------------------------------------
seq2 gattaaagatgttgagttgcatactaagtgggtggtgaggcgagccctcatctgagcccaataacagccaattaccctaattatcaacggcgccccagga
310 320 330 340 350 360 370 380 390 400
seq1 ----------------------------------------------------------------------------------------------------
seq2 cgaggcctagaactgagcgccttgacccaagtcatctcgctccgcctcagcaatgacctgccataaggctcgatcacaccccacttctccaccccgaggc
410 420 430 440 450 460 470 480 490 500
seq1 ----------------------------------------------------------------------------------------------------
seq2 cacggcgtgccgctcacccccgagcctgggtccgggtccggggcccacagctccgccaccatctcggtgtgcagaaactcgaatagcaccgcgtccgcca
510 520 530 540 550 560 570 580 590 600
30 40 50
seq1 --------------------------------------CGGCTTCCATTG-------------------------------GTGTTTTagactagccccg
||||| |||||| ||| | |||| |||
seq2 tacgccgcctcgcgaccctcacgcctggccaacgcttcCGGCTCCCATTGgtcagctcgggaggtaggcccgatctgattgGTGATCCtatttagctccg
610 620 630 640 650 660 670 680 690 700
60 70 80 90 100 110 120 130 140 150
seq1 cctccccgccccggactcaggtggtgtgcgcgtgagcctgggacaCTAGGGGGCGGGCCTACg----CCTTCTCTGGccACATGTGGCAACATCTTCTGC
||| | | ||| |||||| | || || | || | | || ||||||||| ||
seq2 cctgcttcttgaaag------------------------------CCAGGAGGCGGGTCCACcgagcCCCTTCCTTGgcCGCGCTGTTAACATCTTCCGC
710 720 730 740 750 760 770
160 170 180 190 200 210 220 230 240 250
seq1 TCTTACTGCCGtcCCTTCCCTaTTTCCGggttctcgagcATTCCTATTGGCCAAATTACTGGttacacgtgacatactcggcttttaaaatccctattgg
||| || | | || ||||| | ||| || ||| ||| |||||| | | ||| || | | | | | |||| ||
seq2 CCTTTCTTCGCttCCCTCCCTgCTCCCGagtcttcgga-CATCCAATTGGCTGAGTCTCTGAttcctgtgagggtccaagattaaaaaaaataaatttac
780 790 800 810 820 830 840 850 860 87
260 270 280 290 300 310 320
seq1 tccactccagactttcctcgtcgcacaacctggaagagccggcc---------------------------CAGCTGTTTCTCTATTGGCGGTTCCTCCA
| | | || | ||| || | | | | || |||||||| | |||
seq2 gcgtcaaacggaattttcctaaatacataagcaaataaataaataaacccacactacggaagcgacttgtcTAACCGCTTTCCTATTGGCCATAACTCGG
0 880 890 900 910 920 930 940 950 960 97
330 340 350 360 370 380 390 400 410 420
seq1 TTCATGTGACTCTTctgcacgctcactcaaagctgatgctgagaacttgcttcctgattggtagccgggcgaatccacgtgacttggccgctaatgcccg
||| ||||| || | | | ||||
seq2 CTCACGTGACGCTGcgagagaccggctcactt--------------------------------------------------------------------
0 980 990 1000
430 440 450 460 470 480 490 500 510 520
seq1 cccaacccgctcccctattggttgaaatcacagcccttcagctgccacggtgagaacgcagcactcgggttaggaagcggatctcgcaagctccgagcgt
seq2 ----------------------------------------------------------------------------------------------------
530 540 550 560 570 580 590 600 610 620
seq1 cagctgccgggtacggtctttggcgttagcgcttctcccatcccatgagtgcccccagcagagtccagtcggactgtcatcctttctgcgactctggcgc
seq2 ----------------------------------------------------------------------------------------------------
630 640 650 660 670 680 690 700 710 720
seq1 tggtcccgagcagctcacgggcccgtgtgcttccgggtctgagattggcatggtggaccaggtggagggtgtggcctctactaggaggcaaattcgtaaa
seq2 ----------------------------------------------------------------------------------------------------
730 740 750 760 770 780 790 800 810 820
seq1 gacctcggcttgggactccgggaactcgggccccagatccttgttcgagctggtcttcagtttccccatctgtacgctgaagagcctggggtccagtaac
seq2 ----------------------------------------------------------------------------------------------------
830 840 850 860 870 880 890 900 910 920
seq1 ccccatcatcacccagtttacagaagaggaaactgaggcccagacggggagcagctgacaccaagtcgttaagagaatcagcgaaggggctgggaatcca
seq2 ----------------------------------------------------------------------------------------------------
930 940 950 960 970 980 990 1000
seq1 ggacctgcgccttttacccaccgcggcgccggtctcacgtgcagtcccttcgctcttctcccctagttcggtgcc-
seq2 ---------------------------------------------------------------------------C
CONREAL ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00644* | 59.46 | 26 | 32 | 639 | 645 | 1 | 7.026 | 5.546 | 0.87 | 0.80 | Pro only | C only | | MA0095 | 58.33 | 31 | 36 | 644 | 649 | 1 | 6.225 | 5.236 | 0.89 | 0.84 | Yin-Yang | C only | | M00468* | 56.76 | 31 | 37 | 644 | 650 | 2 | 4.840 | 6.111 | 0.81 | 0.88 | Pro only | C only | | M00396 | 59.46 | 38 | 44 | 682 | 688 | 1 | 4.063 | 4.063 | 0.83 | 0.83 | En-1 | C only | | M00469* | 53.85 | 102 | 110 | 716 | 724 | 2 | 4.368 | 6.425 | 0.80 | 0.87 | Pro only | C only | | MA0003 | 53.85 | 102 | 110 | 716 | 724 | 2 | 4.368 | 6.425 | 0.80 | 0.87 | AP2alpha | C only | | M00807* | 53.66 | 103 | 113 | 717 | 727 | 1 | 8.966 | 6.644 | 0.87 | 0.81 | Pro only | C only | | MA0039 | 55.00 | 104 | 113 | 718 | 727 | 1 | 5.228 | 5.483 | 0.81 | 0.82 | Gklf | C only | | M00196 | 53.49 | 104 | 116 | 718 | 730 | 1 | 13.985 | 12.167 | 0.93 | 0.89 | Sp1 | C only | | M00932* | 53.49 | 104 | 116 | 718 | 730 | 1 | 13.863 | 11.412 | 0.93 | 0.88 | Pro only | C only | | M00255 | 52.27 | 104 | 117 | 718 | 731 | 1 | 13.018 | 12.167 | 0.92 | 0.91 | GC box | C only | | M00733* | 51.11 | 104 | 118 | 718 | 732 | 2 | 8.076 | 8.487 | 0.81 | 0.82 | Pro only | C only | | M00716* | 55.26 | 105 | 112 | 719 | 726 | 1 | 7.759 | 5.783 | 0.91 | 0.82 | Pro only | C only | | M00933* | 52.50 | 105 | 114 | 719 | 728 | 2 | 12.418 | 9.829 | 0.94 | 0.87 | Pro only | C only | | M00695* | 54.05 | 106 | 112 | 720 | 726 | 1 | 9.502 | 9.502 | 0.99 | 0.99 | Pro only | C only | | M00649* | 52.63 | 106 | 113 | 720 | 727 | 1 | 10.557 | 6.664 | 0.94 | 0.80 | Pro only | C only | | M00931* | 52.50 | 106 | 115 | 720 | 729 | 1 | 13.288 | 9.813 | 0.95 | 0.87 | Pro only | C only | | M00008 | 52.50 | 106 | 115 | 720 | 729 | 1 | 9.302 | 7.838 | 0.91 | 0.86 | Sp1 | C only | | M00272 | 52.50 | 107 | 116 | 721 | 730 | 1 | 5.226 | 5.534 | 0.81 | 0.82 | p53 | C only | | M00803* | 55.56 | 108 | 113 | 722 | 727 | 1 | 8.243 | 8.243 | 0.97 | 0.97 | Pro only | C only | | M00986* | 55.56 | 110 | 115 | 724 | 729 | 1 | 5.317 | 6.401 | 0.88 | 0.93 | Pro only | C only | | MA0039 | 50.00 | 120 | 129 | 738 | 747 | 2 | 5.047 | 9.038 | 0.81 | 0.92 | Gklf | C only | | M00253 | 57.89 | 132 | 139 | 750 | 757 | 2 | 3.584 | 3.199 | 0.84 | 0.83 | cap | C only | | M00462* | 60.00 | 140 | 149 | 758 | 767 | 2 | 4.504 | 4.431 | 0.80 | 0.80 | Pro only | C only | | M00076 | 60.00 | 140 | 149 | 758 | 767 | 2 | 6.118 | 4.879 | 0.88 | 0.83 | GATA-2 | C only | | M00075 | 60.00 | 140 | 149 | 758 | 767 | 2 | 5.996 | 4.757 | 0.87 | 0.82 | GATA-1 | C only | | MA0037 | 61.11 | 142 | 147 | 760 | 765 | 2 | 3.647 | 3.647 | 0.80 | 0.80 | GATA-3 | C only | | MA0035 | 61.11 | 142 | 147 | 760 | 765 | 2 | 5.784 | 5.784 | 0.93 | 0.93 | GATA-1 | C only | | M00971* | 60.53 | 142 | 149 | 760 | 767 | 1 | 5.302 | 5.302 | 0.80 | 0.80 | Pro only | C only | | M00253 | 60.53 | 142 | 149 | 760 | 767 | 1 | 4.244 | 4.244 | 0.87 | 0.87 | cap | C only | | M00655* | 59.46 | 142 | 148 | 760 | 766 | 1 | 6.158 | 6.158 | 0.84 | 0.84 | Pro only | C only | | MA0036 | 60.00 | 143 | 147 | 761 | 765 | 2 | 5.559 | 5.559 | 0.94 | 0.94 | GATA-2 | C only | | MA0103 | 58.33 | 143 | 148 | 761 | 766 | 1 | 4.388 | 4.388 | 0.81 | 0.81 | deltaEF1 | C only | | M00499* | 60.53 | 144 | 151 | 762 | 769 | 1 | 6.875 | 6.234 | 0.94 | 0.91 | Pro only | C only | | M00496* | 60.53 | 144 | 151 | 762 | 769 | 1 | 4.242 | 5.558 | 0.81 | 0.87 | Pro only | C only | | M00497* | 60.53 | 144 | 151 | 762 | 769 | 1 | 4.422 | 6.144 | 0.83 | 0.91 | Pro only | C only | | MA0080 | 61.11 | 146 | 151 | 764 | 769 | 2 | 5.628 | 8.711 | 0.87 | 1.00 | SPI-1 | C only | | MA0098 | 61.11 | 146 | 151 | 764 | 769 | 1 | 3.822 | 7.797 | 0.82 | 1.00 | c-ETS | C only | | M00658* | 60.53 | 146 | 153 | 764 | 771 | 2 | 7.804 | 7.737 | 0.87 | 0.87 | Pro only | C only | | MA0081 | 59.46 | 147 | 153 | 765 | 771 | 2 | 6.232 | 5.707 | 0.86 | 0.84 | SPI-B | C only | | MA0080 | 69.44 | 154 | 159 | 772 | 777 | 2 | 4.391 | 4.721 | 0.82 | 0.83 | SPI-1 | C only | | MA0080 | 69.44 | 158 | 163 | 776 | 781 | 2 | 4.385 | 4.897 | 0.82 | 0.84 | SPI-1 | C only | | M00649* | 65.79 | 166 | 173 | 784 | 791 | 2 | 7.389 | 11.282 | 0.83 | 0.96 | Pro only | C only | | MA0098 | 63.89 | 168 | 173 | 786 | 791 | 1 | 3.658 | 3.658 | 0.81 | 0.81 | c-ETS | C only | | M00986* | 63.89 | 168 | 173 | 786 | 791 | 2 | 5.392 | 5.768 | 0.88 | 0.90 | Pro only | C only | | MA0098 | 61.11 | 175 | 180 | 793 | 798 | 1 | 7.797 | 3.822 | 1.00 | 0.82 | c-ETS | C only | | MA0098 | 58.33 | 192 | 197 | 809 | 814 | 1 | 5.977 | 5.896 | 0.92 | 0.91 | c-ETS | C only | | M00287 | 54.35 | 192 | 207 | 809 | 824 | 2 | 11.577 | 12.046 | 0.89 | 0.89 | NF-Y | C only | | MA0078 | 56.41 | 195 | 203 | 812 | 820 | 1 | 7.197 | 5.954 | 0.86 | 0.82 | SOX17 | C only | | M00185 | 53.66 | 195 | 205 | 812 | 822 | 2 | 10.383 | 8.635 | 0.90 | 0.85 | NF-Y | C only | | M00254 | 52.38 | 196 | 207 | 813 | 824 | 2 | 11.074 | 8.902 | 0.92 | 0.88 | CCAAT box | C only | | M00775* | 51.16 | 196 | 208 | 813 | 825 | 2 | 10.795 | 10.425 | 0.91 | 0.90 | Pro only | C only | | M00253 | 50.00 | 207 | 214 | 824 | 831 | 2 | 4.068 | 4.193 | 0.86 | 0.87 | cap | C only | | MA0100 | 52.63 | 297 | 304 | 941 | 948 | 2 | 5.172 | 6.947 | 0.81 | 0.86 | c-MYB_1 | C only | | M00497* | 55.26 | 301 | 308 | 945 | 952 | 1 | 5.011 | 4.223 | 0.86 | 0.82 | Pro only | C only | | M00287 | 58.70 | 304 | 319 | 948 | 963 | 2 | 7.875 | 10.407 | 0.82 | 0.86 | NF-Y | C only | | MA0078 | 56.41 | 307 | 315 | 951 | 959 | 1 | 7.197 | 7.197 | 0.86 | 0.86 | SOX17 | C only | | M00395 | 56.41 | 307 | 315 | 951 | 959 | 1 | 3.630 | 5.990 | 0.80 | 0.93 | HOXA3 | C only | | M00775* | 62.79 | 308 | 320 | 952 | 964 | 2 | 7.347 | 8.502 | 0.82 | 0.85 | Pro only | C only | | M00254 | 61.90 | 308 | 319 | 952 | 963 | 2 | 5.836 | 10.535 | 0.81 | 0.91 | CCAAT box | C only | | M00806* | 63.83 | 309 | 325 | 953 | 969 | 2 | 8.622 | 9.290 | 0.82 | 0.84 | Pro only | C only | | MA0109 | 65.79 | 316 | 323 | 960 | 967 | 1 | 4.547 | 5.480 | 0.80 | 0.84 | RUSH1-alfa | C only | | M00985* | 59.09 | 324 | 337 | 968 | 981 | 1 | 8.545 | 13.548 | 0.81 | 0.92 | Pro only | C only | | M00122 | 59.09 | 324 | 337 | 968 | 981 | 1 | 6.810 | 8.849 | 0.83 | 0.88 | USF | C only | | M00122 | 59.09 | 324 | 337 | 968 | 981 | 2 | 6.810 | 8.849 | 0.83 | 0.88 | USF | C only | | MA0067 | 60.53 | 325 | 332 | 969 | 976 | 1 | 5.373 | 4.605 | 0.85 | 0.82 | Pax-2 | C only | | M01008* | 58.97 | 325 | 333 | 969 | 977 | 2 | 6.838 | 9.407 | 0.85 | 0.92 | Pro only | C only | | M00220 | 56.10 | 325 | 335 | 969 | 979 | 1 | 8.439 | 12.721 | 0.81 | 0.92 | SREBP-1 | C only | | M00220 | 56.10 | 326 | 336 | 970 | 980 | 2 | 8.158 | 12.957 | 0.81 | 0.92 | SREBP-1 | C only | | M00187 | 55.00 | 326 | 335 | 970 | 979 | 2 | 6.983 | 10.339 | 0.82 | 0.93 | USF | C only | | MA0093 | 56.76 | 327 | 333 | 971 | 977 | 2 | 7.243 | 10.923 | 0.85 | 0.98 | USF | C only | | M00240 | 56.76 | 327 | 333 | 971 | 977 | 1 | 6.785 | 6.785 | 0.85 | 0.85 | Nkx2-5 | C only | | M01029* | 55.26 | 327 | 334 | 971 | 978 | 1 | 11.893 | 10.351 | 1.00 | 0.95 | Pro only | C only | | M00217 | 55.26 | 327 | 334 | 971 | 978 | 1 | 7.084 | 9.645 | 0.86 | 0.96 | USF | C only | | M00217 | 55.26 | 327 | 334 | 971 | 978 | 2 | 7.967 | 9.645 | 0.89 | 0.96 | USF | C only | | MA0058 | 55.00 | 327 | 336 | 971 | 980 | 2 | 8.345 | 9.621 | 0.86 | 0.89 | Max | C only | | MA0004 | 55.56 | 328 | 333 | 972 | 977 | 1 | 6.112 | 10.351 | 0.83 | 1.00 | ARNT | C only | | MA0004 | 55.56 | 328 | 333 | 972 | 977 | 2 | 6.112 | 10.351 | 0.83 | 1.00 | ARNT | C only | | MA0104 | 55.56 | 328 | 333 | 972 | 977 | 1 | 6.819 | 10.533 | 0.86 | 1.00 | n-MYC | C only | | MA0104 | 55.56 | 328 | 333 | 972 | 977 | 2 | 7.418 | 10.533 | 0.88 | 1.00 | n-MYC | C only | | M00726* | 55.56 | 328 | 333 | 972 | 977 | 1 | 6.970 | 8.133 | 0.94 | 1.00 | Pro only | C only | | M00726* | 55.56 | 328 | 333 | 972 | 977 | 2 | 4.701 | 8.133 | 0.82 | 1.00 | Pro only | C only | | MA0103 | 55.56 | 328 | 333 | 972 | 977 | 2 | 4.633 | 6.156 | 0.82 | 0.88 | deltaEF1 | C only | | MA0093 | 54.05 | 328 | 334 | 972 | 978 | 1 | 7.800 | 10.923 | 0.87 | 0.98 | USF | C only | | M01008* | 53.85 | 328 | 336 | 972 | 980 | 1 | 7.495 | 10.853 | 0.87 | 0.97 | Pro only | C only | | MA0067 | 52.63 | 329 | 336 | 973 | 980 | 2 | 6.078 | 8.225 | 0.88 | 0.97 | Pax-2 | C only | | MA0067 | 52.63 | 330 | 337 | 974 | 981 | 1 | 4.474 | 5.006 | 0.82 | 0.84 | Pax-2 | C only | | MA0089 | 50.00 | 330 | 335 | 974 | 979 | 1 | 5.300 | 6.040 | 0.85 | 0.88 | TCF11-MafG | C only | | M00925* | 53.85 | 331 | 339 | 975 | 983 | 1 | 7.949 | 5.695 | 0.88 | 0.80 | Pro only | C only | | M00801* | 50.00 | 331 | 336 | 975 | 980 | 2 | 5.524 | 9.044 | 0.85 | 0.99 | Pro only | C only |
LAGAN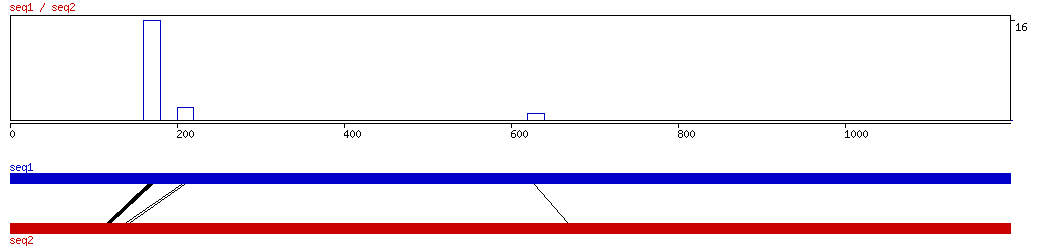
LAGAN ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 cgtcatggcgcgagccctcctactccggcttccattggtgttttagactagccccgcctccccgccccggactcaggtggtgtgcgcgtgagcctgggac
| | | || | | | | ||| | ||| || ||
seq2 ggatgagaaggaagaca---------------------------------------------------------atgggaggtggatgggagtatgacac
10 20 30 40
110 120 130 140 150 160 170 180
seq1 actagggggcgggcctacgccttctctggccacatgtggcaacatct------------------tctgctcttactgccgtCCCTTCCCTATTTccggg
|| | ||| | || | |||| | || || | | | || |||||| ||| |||||
seq2 acaggaaggcacaaaggaaca---------cataagtggtggtaactattattgattacaatatttcagtgatcccagcgccACCTTCCACATTAccggg
50 60 70 80 90 100 110 120 130
190 200 210 220 230 240 250 260
seq1 ttctcgagcattcctattggccAAATTACTGGTTacacgtgacatactcggcttttaaaatccctattggtc---------------cactccagacttt
| || ||||| ||||| | | | | || ||| || || | | || | |||||
seq2 t-----------------ggggTCTTTACTCATTacaggggcaaatccatactaaaaaactcactgtgtgctggggtgtagcgcgatcagcacgcacttt
140 150 160 170 180 190 200 210
270 280 290 300 310 320 330
seq1 cctcgtcgcacaacctggaagagccggcccagctgtttctctattggcggttcctccattcatgtga---------------------------------
| | |||| | | | | | | | | | ||| ||| | | | | | |
seq2 atccagcacacaggcagaggggacagatgagaccgggtatg-gttgagggtgcattttctaaggaggaaacggaaggataggaagattaaagatgttgag
220 230 240 250 260 270 280 290 300 310
340 350 360 370 380 390 400
seq1 ----------------------------ctcttctgcacgctcactcaaagctgatgctgagaacttgcttcctgattggtagccgggcgaatccacgtg
| || | || || | | ||| || | | | || | | || || | |
seq2 ttgcatactaagtgggtggtgaggcgagccctcatctgagcccaataacagccaatt-----accctaattatcaacggcgccccaggacgaggcctaga
320 330 340 350 360 370 380 390 400 410
410 420 430 440 450 460 470
seq1 acttggccgctaatgcccgcccaacccgctcccct----------------attggttgaaatcacagccc---------ttcagctgccacggtgag--
||| || || || || | |||||| | || | |||||| ||| | | ||||||| | |
seq2 actgagcgccttgacccaagtcatctcgctccgcctcagcaatgacctgccataaggctcgatcacaccccacttctccaccccgaggccacggcgtgcc
420 430 440 450 460 470 480 490 500 510
480 490 500 510 520 530 540 550 560 570
seq1 ----aacgcagcactcgggttaggaagcggatctcgcaagctccgagcgtcagctgccgggtacggtctttggcgttagcgcttctcccatcccatgagt
| | | | | |||| || ||| | | || |||||| | | | | || | ||| || |||| | | |
seq2 gctcacccccgagcctgggtccgggtccggggcccaca-gctccgccaccatctcggtgtgcagaaactcgaatagcaccgcgtccgccatacgccgcct
520 530 540 550 560 570 580 590 600 610
580 590 600 610 620 630 640 650 660
seq1 ----gcccccagcagagtccagtcggactgtcatcctttctgcgactctggcgctgGTCCCGagcagctca-----------cgggcccgtgtgcttccg
||| || | ||| | | | || || | ||| || | | | ||||| | | | | ||| ||||||
seq2 cgcgaccctcacgcctggccaacgcttccggctcccattggtcagctcgggaggtaGGCCCGatctgattggtgatcctatttagctccgcctgcttctt
620 630 640 650 660 670 680 690 700 710
670 680 690 700 710 720 730 740 750
seq1 ggtctgagattggcatggtggaccaggtggagggtgtggcctctactaggaggcaaattcgtaaagacctcggcttgggactccgggaa-----ctcggg
| | ||||||| | | || || ||||| | | | || |
seq2 gaaa----------------------------------------gccaggaggcgggtccaccgagccccttccttggccgcgctgttaacatcttccgc
720 730 740 750 760 770
760 770 780 790 800 810 820 830 840 850
seq1 ccccagatccttgttcgagctggtcttcagtttccccatctgtacgctgaagagcctggggtccagtaacccccatcatcacccagtttacagaagagga
|| || | | ||| || ||| | | | | | | ||| || | | || | | ||| | | || | |
seq2 cctttcttcgcttccctccctgctcccgagtcttcggacatccaattggctgagtctc--------tgattcctgtgagggtccaagattaaaaaaaata
780 790 800 810 820 830 840 850 860
860 870 880 890 900 910 920 930 940
seq1 aactgaggcccagacggggagcagctgacaccaagtcgttaagagaatcagcgaaggggctgggaatccaggacctgcgccttttacc------------
|| | | || | || | | | || | |||| ||| | || | | | | | | | | | | |||
seq2 aatttacgcgtcaaacggaattttc-----ctaaatacataagcaaataaataaataaacccacactacggaagcgacttgtctaaccgctttcctattg
870 880 890 900 910 920 930 940 950
950 960 970 980 990 1000
seq1 ---------------------caccgcg-gcgccggtctcacgt----------gcagtcccttcgctcttctcccctag--------ttcggtgcc
| | ||| | | | | ||||| | || | ||| | | || ||| | || |||||
seq2 gccataactcggctcacgtgacgctgcgagagaccggctcacttccgccaggcagcggaccccttgggctcggagcctcggagacccgctccgtgcc
960 970 980 990 1000 1010 1020 1030 1040 1050
LAGAN ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00496* | 52.63 | 165 | 172 | 117 | 124 | 1 | 5.438 | 4.321 | 0.86 | 0.82 | Pro only | L M | | M00497* | 52.63 | 165 | 172 | 117 | 124 | 1 | 7.250 | 5.348 | 0.96 | 0.87 | Pro only | L M | | M00499* | 52.63 | 165 | 172 | 117 | 124 | 1 | 5.727 | 5.596 | 0.89 | 0.88 | Pro only | L M | | M00500* | 52.63 | 165 | 172 | 117 | 124 | 1 | 8.262 | 7.040 | 0.96 | 0.91 | Pro only | L M | | M00704* | 55.56 | 166 | 171 | 118 | 123 | 2 | 5.117 | 5.117 | 0.82 | 0.82 | Pro only | L M | | M00971* | 52.63 | 166 | 173 | 118 | 125 | 1 | 6.109 | 6.702 | 0.83 | 0.85 | Pro only | L M | | MA0039 | 50.00 | 166 | 175 | 118 | 127 | 2 | 8.425 | 6.082 | 0.90 | 0.84 | Gklf | L M | | M00658* | 50.00 | 167 | 174 | 119 | 126 | 2 | 6.103 | 7.704 | 0.81 | 0.87 | Pro only | L M | | MA0080 | 52.78 | 167 | 172 | 119 | 124 | 2 | 8.579 | 6.483 | 0.99 | 0.91 | SPI-1 | L M | | MA0098 | 52.78 | 167 | 172 | 119 | 124 | 1 | 5.777 | 6.300 | 0.91 | 0.93 | c-ETS | L M | | MA0081 | 51.35 | 168 | 174 | 120 | 126 | 2 | 5.764 | 6.960 | 0.84 | 0.88 | SPI-B | L M | | M00690* | 50.00 | 169 | 176 | 121 | 128 | 1 | 5.863 | 8.905 | 0.85 | 0.97 | Pro only | L M | | MA0056 | 52.78 | 169 | 174 | 121 | 126 | 2 | 4.916 | 4.989 | 0.81 | 0.81 | MZF_1-4 | L M | | M00313* | 50.00 | 170 | 177 | 122 | 129 | 1 | 4.491 | 4.766 | 0.81 | 0.82 | Pro only | L M | | M00314* | 50.00 | 170 | 177 | 122 | 129 | 1 | 4.587 | 4.950 | 0.81 | 0.83 | Pro only | L M | | M00315* | 50.00 | 170 | 177 | 122 | 129 | 1 | 4.418 | 4.516 | 0.81 | 0.81 | Pro only | L M | | M00496* | 50.00 | 205 | 212 | 140 | 147 | 1 | 4.409 | 4.162 | 0.82 | 0.81 | Pro only | L M | | MA0032 | 50.00 | 209 | 216 | 144 | 151 | 2 | 5.238 | 5.764 | 0.88 | 0.90 | FREAC-3 | L M | | M00986* | 52.78 | 628 | 633 | 667 | 672 | 2 | 6.604 | 5.317 | 0.94 | 0.88 | Pro only | L only |
MAVID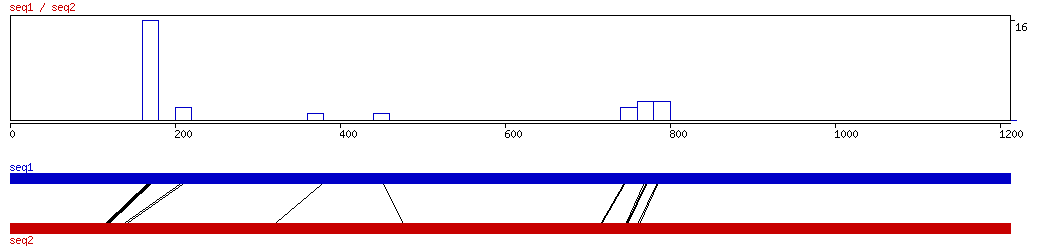
MAVID ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 cgtcatggcgcgagccctcctactccggcttccattggtgttttagactagccccgcctccccgccccggactcaggtggtgtgcgcgtgagcctgggac
| | | || | | | | ||| | ||| || ||
seq2 ggatgagaaggaagac---------------------------------------------------------aatgggaggtggatgggagtatgacac
10 20 30 40
110 120 130 140 150 160 170 180
seq1 actagggggcgggcctacgccttctctggccacat-----gtggcaacatcttctgctcttact---------------gccgtCCCTTCCCTATTTccg
|| || || | | | | ||||| |||| ||| | || | | | || |||||| ||| |||
seq2 ac---aggaaggcacaaag--------gaacacataagtggtggtaactattattgattacaatatttcagtgatcccagcgccACCTTCCACATTAccg
50 60 70 80 90 100 110 120 130
190 200 210 220 230 240 250 260 27
seq1 ggttctcgagcattcctattggccAAATTACTGGTTacacgtgacatactcggcttttaaaatccctat-----------tggtccactccagactttcc
|| ||| ||||| ||||| | | | | || ||| || || | | | | || || |
seq2 gg-----------------tggggTCTTTACTCATTacaggggcaaatccatactaaaaaactcactgtgtgctggggtgtagcgcgatcagcacgcact
140 150 160 170 180 190 200 210
0 280 290 300 310 320 330 340 350 360
seq1 tcgtcgcacaacctggaagagccggcccagctg--------tttctctattggcggttcctccattcatgtgactcttctgcacgctcactcaaagctga
| || || | || |||| || ||| || | | | || | | || | | | || | | | | || ||
seq2 ttatccagcacacaggcagag--gggacagatgagaccgggtatggttgagggtgcattttctaaggaggaaacggaaggataggaagattaaa----ga
220 230 240 250 260 270 280 290 300 31
370 380 390 400 410
seq1 tgctgagaacttgcttCCTGATTGGtagccg-ggcgaatccacgt-----------------------------------------------gacttggc
|| |||| |||| | || | ||| | | ||||| || | | ||| |||
seq2 tgttgag---ttgcatACTAAGTGGgtggtgaggcgagccctcatctgagcccaataacagccaattaccctaattatcaacggcgccccaggacgaggc
0 320 330 340 350 360 370 380 390 400
420 430 440 450 460 470 480
seq1 cgctaa-tgcccgcc-----------caacccgctcccc------------------tattggttgaaaTCACAGCCcttcagctgccacggtgagaacg
| || || |||| || | |||||| | || | | || ||| | |||| |||| ||| |
seq2 ctagaactgagcgccttgacccaagtcatctcgctccgcctcagcaatgacctgccataaggctcgatcACACCCCActtc----tccaccccgaggcca
410 420 430 440 450 460 470 480 490 500
490 500 510 520 530 540 550 560 570 580
seq1 cagcactcgggttaggaagcggatctcgcaagctccgagcgtcagctgccgggtacggtctttggcgttagcgcttctcccatcccatgagtgcccccag
| || | | | | | |||||| | | |||||| ||| | | ||| | | ||||| | | ||||
seq2 cggcgtgccgct-----------------cacccccgagc--ctgggtccgggtccgg----ggcccacagctccgccaccatctc-ggtgtgc-----a
510 520 530 540 550 560 570
590 600 610 620 630 640 650 660 670 680
seq1 cagagtccagtcggactg--tcatcctttctgcgactctggcgctggtcccgagcagctcacgggcccgtgtgcttccgggtctgagattggcatggtgg
| | || | | | || | || || | | || ||| ||| || || || || |||||||| || | | |
seq2 gaaactcgaatagcaccgcgtccgccatacgccgcctc--gcgaccctcacgcctggccaac----------gcttccggctccca-----------ttg
580 590 600 610 620 630 640 650
690 700 710 720 730 740 750 760
seq1 accaggtggagggtgtggcc---tctactaggaggcaaattcgtaaagacctcggcttgggact------cCGGGAACTcgggccccaga------tcct
||| | | | | |||| ||| | || | || || | ||| || || || || ||| || || | || ||||
seq2 gtcagctcgggaggtaggcccgatctgattggtg-----atcctatttagctccgcctgcttcttgaaagcCAGGAGGCgggtccaccgagccccttcct
660 670 680 690 700 710 720 730 740
770 780 790 800 810 820 830
seq1 tGTTCGAGCTGGTcttCAGTTTCCccatctgtacgctgaagagcctggggtccagtaacccccatcatca------------------------------
|| || |||| | || | || | | | |||| ||| | ||| | | | ||||
seq2 tGGCCGCGCTGTTaacATCTTCCGccctttcttcgcttccctccct--gctcccgagtcttcggacatccaattggctgagtctctgattcctgtgaggg
750 760 770 780 790 800 810 820 830 840
840 850 860 870 880 890 900 910 920
seq1 -cccagtttacagaagaggaaactgaggcccagacggggagcagctgac------accaagtcgttaagagaatcagcgaaggggctgggaat-------
|| || ||| | || | || | | || |||| || | | ||| | ||| || | | || | || |
seq2 tccaagattaaaaaaaataaatttacgcgtcaaacggaattttcctaaatacataagcaaataaataaataaacccacactacggaagcgacttgtctaa
850 860 870 880 890 900 910 920 930 940
930 940 950 960 970 980 990
seq1 ---------------ccaggac-ctgcgccttttacccaccgcggcgccggtctcacgt----------gcagtcccttcgctcttctcccctagt----
||| || | || | | || | || | |||| ||||| | || | ||| | | || ||| |
seq2 ccgctttcctattggccataactcggctcacgtgacgctgcgagagaccgg-ctcacttccgccaggcagcggaccccttgggctcggagcctcggagac
950 960 970 980 990 1000 1010 1020 1030 1040
1000
seq1 ----tcggtgcc
|| |||||
seq2 ccgctccgtgcc
1050
MAVID ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00496* | 52.63 | 165 | 172 | 117 | 124 | 1 | 5.438 | 4.321 | 0.86 | 0.82 | Pro only | M L | | M00497* | 52.63 | 165 | 172 | 117 | 124 | 1 | 7.250 | 5.348 | 0.96 | 0.87 | Pro only | M L | | M00499* | 52.63 | 165 | 172 | 117 | 124 | 1 | 5.727 | 5.596 | 0.89 | 0.88 | Pro only | M L | | M00500* | 52.63 | 165 | 172 | 117 | 124 | 1 | 8.262 | 7.040 | 0.96 | 0.91 | Pro only | M L | | M00704* | 55.56 | 166 | 171 | 118 | 123 | 2 | 5.117 | 5.117 | 0.82 | 0.82 | Pro only | M L | | M00971* | 52.63 | 166 | 173 | 118 | 125 | 1 | 6.109 | 6.702 | 0.83 | 0.85 | Pro only | M L | | MA0039 | 50.00 | 166 | 175 | 118 | 127 | 2 | 8.425 | 6.082 | 0.90 | 0.84 | Gklf | M L | | M00658* | 50.00 | 167 | 174 | 119 | 126 | 2 | 6.103 | 7.704 | 0.81 | 0.87 | Pro only | M L | | MA0080 | 52.78 | 167 | 172 | 119 | 124 | 2 | 8.579 | 6.483 | 0.99 | 0.91 | SPI-1 | M L | | MA0098 | 52.78 | 167 | 172 | 119 | 124 | 1 | 5.777 | 6.300 | 0.91 | 0.93 | c-ETS | M L | | MA0081 | 51.35 | 168 | 174 | 120 | 126 | 2 | 5.764 | 6.960 | 0.84 | 0.88 | SPI-B | M L | | M00690* | 50.00 | 169 | 176 | 121 | 128 | 1 | 5.863 | 8.905 | 0.85 | 0.97 | Pro only | M L | | MA0056 | 52.78 | 169 | 174 | 121 | 126 | 2 | 4.916 | 4.989 | 0.81 | 0.81 | MZF_1-4 | M L | | M00313* | 50.00 | 170 | 177 | 122 | 129 | 1 | 4.491 | 4.766 | 0.81 | 0.82 | Pro only | M L | | M00314* | 50.00 | 170 | 177 | 122 | 129 | 1 | 4.587 | 4.950 | 0.81 | 0.83 | Pro only | M L | | M00315* | 50.00 | 170 | 177 | 122 | 129 | 1 | 4.418 | 4.516 | 0.81 | 0.81 | Pro only | M L | | M00496* | 50.00 | 205 | 212 | 140 | 147 | 1 | 4.409 | 4.162 | 0.82 | 0.81 | Pro only | M L | | MA0032 | 50.00 | 209 | 216 | 144 | 151 | 2 | 5.238 | 5.764 | 0.88 | 0.90 | FREAC-3 | M L | | M00395 | 53.85 | 378 | 386 | 323 | 331 | 1 | 5.580 | 3.968 | 0.91 | 0.82 | HOXA3 | M only | | M00253 | 52.63 | 453 | 460 | 476 | 483 | 1 | 4.554 | 3.833 | 0.88 | 0.85 | cap | M only | | M00971* | 50.00 | 744 | 751 | 717 | 724 | 2 | 5.951 | 6.429 | 0.82 | 0.84 | Pro only | M only | | MA0098 | 50.00 | 745 | 750 | 718 | 723 | 2 | 4.122 | 3.674 | 0.83 | 0.81 | c-ETS | M only | | M00428* | 50.00 | 768 | 775 | 747 | 754 | 1 | 4.677 | 4.130 | 0.84 | 0.81 | Pro only | M only | | M00175 | 55.00 | 770 | 779 | 749 | 758 | 2 | 6.386 | 7.305 | 0.81 | 0.85 | AP-4 | M only | | M00253 | 52.63 | 771 | 778 | 750 | 757 | 2 | 3.772 | 3.199 | 0.85 | 0.83 | cap | M only | | M00492* | 52.63 | 783 | 790 | 762 | 769 | 1 | 5.424 | 5.442 | 0.80 | 0.80 | Pro only | M only | | M00497* | 52.63 | 783 | 790 | 762 | 769 | 1 | 3.745 | 6.144 | 0.80 | 0.91 | Pro only | M only | | MA0080 | 52.78 | 785 | 790 | 764 | 769 | 2 | 4.264 | 8.711 | 0.81 | 1.00 | SPI-1 | M only |
BLASTZ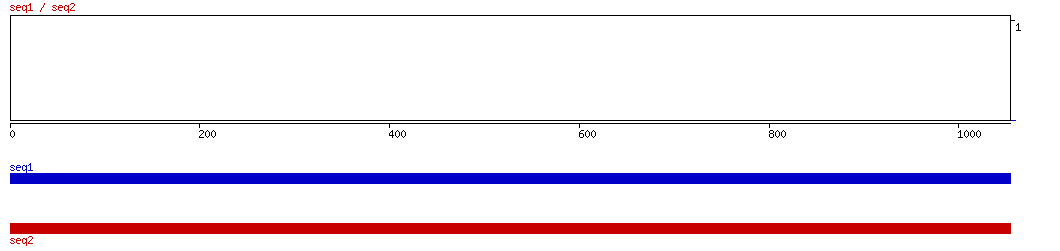
BLASTZ ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 cgtcatggcgcgagccctcctactccggcttccattggtgttttagactagccccgcctccccgccccggactcaggtggtgtgcgcgtgagcctgggac
| | | || | || | | | || | | | | | | | | | |
seq2 ggatgagaaggaagacaatgggaggtggatgggagtatgacacacaggaaggcacaaaggaacacataagtggtggtaactattattgattacaatattt
10 20 30 40 50 60 70 80 90 100
110 120 130 140 150 160 170 180 190 200
seq1 actagggggcgggcctacgccttctctggccacatgtggcaacatcttctgctcttactgccgtcccttccctatttccgggttctcgagcattcctatt
| | | | || | || || || || | | | | | | | |
seq2 cagtgatcccagcgccaccttccacattaccgggtggggtctttactcattacaggggcaaatccatactaaaaaactcactgtgtgctggggtgtagcg
110 120 130 140 150 160 170 180 190 200
210 220 230 240 250 260 270 280 290 300
seq1 ggccaaattactggttacacgtgacatactcggcttttaaaatccctattggtccactccagactttcctcgtcgcacaacctggaagagccggcccagc
| | || | | | | | | || | | | | | | |
seq2 cgatcagcacgcactttatccagcacacaggcagaggggacagatgagaccgggtatggttgagggtgcattttctaaggaggaaacggaaggataggaa
210 220 230 240 250 260 270 280 290 300
310 320 330 340 350 360 370 380 390 400
seq1 tgtttctctattggcggttcctccattcatgtgactcttctgcacgctcactcaaagctgatgctgagaacttgcttcctgattggtagccgggcgaatc
|| | | | | | | | | | | | |||| | | || | || | | | || | |
seq2 gattaaagatgttgagttgcatactaagtgggtggtgaggcgagccctcatctgagcccaataacagccaattaccctaattatcaacggcgccccagga
310 320 330 340 350 360 370 380 390 400
410 420 430 440 450 460 470 480 490 500
seq1 cacgtgacttggccgctaatgcccgcccaacccgctcccctattggttgaaatcacagcccttcagctgccacggtgagaacgcagcactcgggttagga
| | | | ||||| | | || | | | | | | | | | | |||
seq2 cgaggcctagaactgagcgccttgacccaagtcatctcgctccgcctcagcaatgacctgccataaggctcgatcacaccccacttctccaccccgaggc
410 420 430 440 450 460 470 480 490 500
510 520 530 540 550 560 570 580 590 600
seq1 agcggatctcgcaagctccgagcgtcagctgccgggtacggtctttggcgttagcgcttctcccatcccatgagtgcccccagcagagtccagtcggact
||| | || | | | | |||||| ||| | | | | | || | | | || | ||| | |
seq2 cacggcgtgccgctcacccccgagcctgggtccgggtccggggcccacagctccgccaccatctcggtgtgcagaaactcgaatagcaccgcgtccgcca
510 520 530 540 550 560 570 580 590 600
610 620 630 640 650 660 670 680 690 700
seq1 gtcatcctttctgcgactctggcgctggtcccgagcagctcacgggcccgtgtgcttccgggtctgagattggcatggtggaccaggtggagggtgtggc
| | || | |||| | | | || || | | | || | | | |
seq2 tacgccgcctcgcgaccctcacgcctggccaacgcttccggctcccattggtcagctcgggaggtaggcccgatctgattggtgatcctatttagctccg
610 620 630 640 650 660 670 680 690 700
710 720 730 740 750 760 770 780 790 800
seq1 ctctactaggaggcaaattcgtaaagacctcggcttgggactccgggaactcgggccccagatccttgttcgagctggtcttcagtttccccatctgtac
| | | | ||| | | || | ||| | | | | || || |
seq2 cctgcttcttgaaagccaggaggcgggtccaccgagccccttccttggccgcgctgttaacatcttccgccctttcttcgcttccctccctgctcccgag
710 720 730 740 750 760 770 780 790 800
810 820 830 840 850 860 870 880 890 900
seq1 gctgaagagcctggggtccagtaacccccatcatcacccagtttacagaagaggaaactgaggcccagacggggagcagctgacaccaagtcgttaagag
|| || | | | | | || | || | ||| | | | | | ||
seq2 tcttcggacatccaattggctgagtctctgattcctgtgagggtccaagattaaaaaaaataaatttacgcgtcaaacggaattttcctaaatacataag
810 820 830 840 850 860 870 880 890 900
910 920 930 940 950 960 970 980 990 1000
seq1 aatcagcgaaggggctgggaatccaggacctgcgccttttacccaccgcggcgccggtctcacgtgcagtcccttcgctcttctcccctagttcggtgcc
| | | | | | | | | | | | | | | | | | | | |
seq2 caaataaataaataaacccacactacggaagcgacttgtctaaccgctttcctattggccataactcggctcacgtgacgctgcgagagaccggctcact
910 920 930 940 950 960 970 980 990 1000
seq1 ------------------------------------------------------
seq2 tccgccaggcagcggaccccttgggctcggagcctcggagacccgctccgtgcc
1010 1020 1030 1040 1050
BLASTZ ALIGNED HITSNo hits found
Run time: 86.9 sec | 




