GENOMIC
Mapping
10qA3. View the map and BAC clones (data from UCSC genome browser).
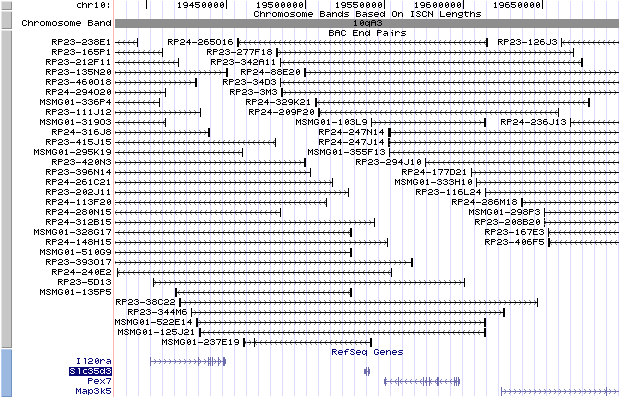
A BAC
(RP23-344M6) containing the wild-type copy of the Slc35d3 gene was inserted
into fertilized eggs and rescued the Slc35d3-/- phenotype (Chintala, et al).
Structure
(assembly 02/2006)
Slc35d3/NM_029529: 2 exons, 3,540 bp, chr10:19,537,333-19,540,872.
The figure below shows the structure of the Slc35d3 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Slc35d3, seq2=human SLC35D3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Slc35d3 ( NM_029529), 2,630 bp, view ORF and the alignment to genomic.
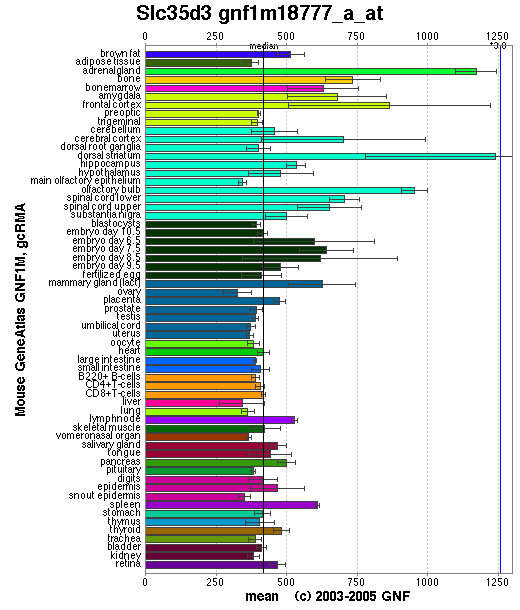
Expression Pattern
Tissue specificity: mainly expressed in brain. Significant expression of Slc35d3
was observed in bone marrow and platelets (Chintala, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
frc, fringe-like 1 (NP_083805): 422 aa, UniProtKB/Swiss-Prot entry Q8BGF8.
Ortholog
Species
Human Chimpanzee Rat Dog Fowl GeneView
SLC35D3
SLC35D3
Slc35d3
LOC484001
LOC428614
Protein
NP_001008783 (416aa)
XP_518764 (416aa)
XP_218777 (420aa)
XP_541118 (423aa)
XP_426171 (599aa)
Identities
380/422 (90%) 380/422 (90%) 409/422 (96%) 372/423 (87%) 176/275 (64%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
TPT domain: 157-299.
UAA domain: 13-305.
(2) Transmembrane domains predicted by HMMTOP:
N-terminus: IN
Number of transmembrane helices: 10
Transmembrane helices: 9-29, 38-56, 63-87, 104-123, 130-149, 156-175, 188-207, 224-243, 252-271, 280-299.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
10qA3. View the map and BAC clones (data from UCSC genome browser).

A BAC (RP23-344M6) containing the wild-type copy of the Slc35d3 gene was inserted into fertilized eggs and rescued the Slc35d3-/- phenotype (Chintala, et al).
Structure
(assembly 02/2006)
Slc35d3/NM_029529: 2 exons, 3,540 bp, chr10:19,537,333-19,540,872.
The figure below shows the structure of the Slc35d3 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Slc35d3, seq2=human SLC35D3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Slc35d3 ( NM_029529), 2,630 bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: mainly expressed in brain. Significant expression of Slc35d3
was observed in bone marrow and platelets (Chintala, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.

PROTEIN
Sequence
frc, fringe-like 1 (NP_083805): 422 aa, UniProtKB/Swiss-Prot entry Q8BGF8.
Ortholog
Species
Human Chimpanzee Rat Dog Fowl GeneView
SLC35D3
SLC35D3
Slc35d3
LOC484001
LOC428614
Protein
NP_001008783 (416aa)
XP_518764 (416aa)
XP_218777 (420aa)
XP_541118 (423aa)
XP_426171 (599aa)
Identities
380/422 (90%) 380/422 (90%) 409/422 (96%) 372/423 (87%) 176/275 (64%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
TPT domain: 157-299.
UAA domain: 13-305.
(2) Transmembrane domains predicted by HMMTOP:
N-terminus: IN
Number of transmembrane helices: 10
Transmembrane helices: 9-29, 38-56, 63-87, 104-123, 130-149, 156-175, 188-207, 224-243, 252-271, 280-299.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
(assembly 02/2006)
Slc35d3/NM_029529: 2 exons, 3,540 bp, chr10:19,537,333-19,540,872.
The figure below shows the structure of the Slc35d3 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Slc35d3, seq2=human SLC35D3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Slc35d3 ( NM_029529), 2,630 bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: mainly expressed in brain. Significant expression of Slc35d3
was observed in bone marrow and platelets (Chintala, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.

PROTEIN
Sequence
frc, fringe-like 1 (NP_083805): 422 aa, UniProtKB/Swiss-Prot entry Q8BGF8.
Ortholog
Species
Human Chimpanzee Rat Dog Fowl GeneView
SLC35D3
SLC35D3
Slc35d3
LOC484001
LOC428614
Protein
NP_001008783 (416aa)
XP_518764 (416aa)
XP_218777 (420aa)
XP_541118 (423aa)
XP_426171 (599aa)
Identities
380/422 (90%) 380/422 (90%) 409/422 (96%) 372/423 (87%) 176/275 (64%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
TPT domain: 157-299.
UAA domain: 13-305.
(2) Transmembrane domains predicted by HMMTOP:
N-terminus: IN
Number of transmembrane helices: 10
Transmembrane helices: 9-29, 38-56, 63-87, 104-123, 130-149, 156-175, 188-207, 224-243, 252-271, 280-299.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
Search the 5'UTR and 1kb upstream regions (seq1=mouse Slc35d3, seq2=human SLC35D3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Slc35d3 ( NM_029529), 2,630 bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: mainly expressed in brain. Significant expression of Slc35d3
was observed in bone marrow and platelets (Chintala, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.

PROTEIN
Sequence
frc, fringe-like 1 (NP_083805): 422 aa, UniProtKB/Swiss-Prot entry Q8BGF8.
Ortholog
Species
Human Chimpanzee Rat Dog Fowl GeneView
SLC35D3
SLC35D3
Slc35d3
LOC484001
LOC428614
Protein
NP_001008783 (416aa)
XP_518764 (416aa)
XP_218777 (420aa)
XP_541118 (423aa)
XP_426171 (599aa)
Identities
380/422 (90%) 380/422 (90%) 409/422 (96%) 372/423 (87%) 176/275 (64%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
TPT domain: 157-299.
UAA domain: 13-305.
(2) Transmembrane domains predicted by HMMTOP:
N-terminus: IN
Number of transmembrane helices: 10
Transmembrane helices: 9-29, 38-56, 63-87, 104-123, 130-149, 156-175, 188-207, 224-243, 252-271, 280-299.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
Slc35d3 ( NM_029529), 2,630 bp, view ORF and the alignment to genomic.
Expression Pattern
Tissue specificity: mainly expressed in brain. Significant expression of Slc35d3
was observed in bone marrow and platelets (Chintala, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.

PROTEIN
Sequence
frc, fringe-like 1 (NP_083805): 422 aa, UniProtKB/Swiss-Prot entry Q8BGF8.
Ortholog
Species
Human Chimpanzee Rat Dog Fowl GeneView
SLC35D3
SLC35D3
Slc35d3
LOC484001
LOC428614
Protein
NP_001008783 (416aa)
XP_518764 (416aa)
XP_218777 (420aa)
XP_541118 (423aa)
XP_426171 (599aa)
Identities
380/422 (90%) 380/422 (90%) 409/422 (96%) 372/423 (87%) 176/275 (64%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
TPT domain: 157-299.
UAA domain: 13-305.
(2) Transmembrane domains predicted by HMMTOP:
N-terminus: IN
Number of transmembrane helices: 10
Transmembrane helices: 9-29, 38-56, 63-87, 104-123, 130-149, 156-175, 188-207, 224-243, 252-271, 280-299.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
Tissue specificity: mainly expressed in brain. Significant expression of Slc35d3 was observed in bone marrow and platelets (Chintala, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.

PROTEIN
Sequence
frc, fringe-like 1 (NP_083805): 422 aa, UniProtKB/Swiss-Prot entry Q8BGF8.
Ortholog
Species
Human Chimpanzee Rat Dog Fowl GeneView
SLC35D3
SLC35D3
Slc35d3
LOC484001
LOC428614
Protein
NP_001008783 (416aa)
XP_518764 (416aa)
XP_218777 (420aa)
XP_541118 (423aa)
XP_426171 (599aa)
Identities
380/422 (90%) 380/422 (90%) 409/422 (96%) 372/423 (87%) 176/275 (64%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
TPT domain: 157-299.
UAA domain: 13-305.
(2) Transmembrane domains predicted by HMMTOP:
N-terminus: IN
Number of transmembrane helices: 10
Transmembrane helices: 9-29, 38-56, 63-87, 104-123, 130-149, 156-175, 188-207, 224-243, 252-271, 280-299.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
frc, fringe-like 1 (NP_083805): 422 aa, UniProtKB/Swiss-Prot entry Q8BGF8.
Ortholog
Species
Human Chimpanzee Rat Dog Fowl GeneView
SLC35D3
SLC35D3
Slc35d3
LOC484001
LOC428614
Protein
NP_001008783 (416aa)
XP_518764 (416aa)
XP_218777 (420aa)
XP_541118 (423aa)
XP_426171 (599aa)
Identities
380/422 (90%) 380/422 (90%) 409/422 (96%) 372/423 (87%) 176/275 (64%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
TPT domain: 157-299.
UAA domain: 13-305.
(2) Transmembrane domains predicted by HMMTOP:
N-terminus: IN
Number of transmembrane helices: 10
Transmembrane helices: 9-29, 38-56, 63-87, 104-123, 130-149, 156-175, 188-207, 224-243, 252-271, 280-299.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
| Species | Human | Chimpanzee | Rat | Dog | Fowl |
| GeneView | SLC35D3 | SLC35D3 | Slc35d3 | LOC484001 | LOC428614 |
| Protein | NP_001008783 (416aa) | XP_518764 (416aa) | XP_218777 (420aa) | XP_541118 (423aa) | XP_426171 (599aa) |
| Identities | 380/422 (90%) | 380/422 (90%) | 409/422 (96%) | 372/423 (87%) | 176/275 (64%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
TPT domain: 157-299.
UAA domain: 13-305.
(2) Transmembrane domains predicted by HMMTOP:
N-terminus: IN
Number of transmembrane helices: 10
Transmembrane helices: 9-29, 38-56, 63-87, 104-123, 130-149, 156-175, 188-207, 224-243, 252-271, 280-299.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
(1) Domains predicted by SMART:
TPT domain: 157-299.
UAA domain: 13-305.
(2) Transmembrane domains predicted by HMMTOP:
N-terminus: IN
Number of transmembrane helices: 10
Transmembrane helices: 9-29, 38-56, 63-87, 104-123, 130-149, 156-175, 188-207, 224-243, 252-271, 280-299.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
(1) Predicted results by ScanProsite:
a) N-glycosylation site: 240 to 243 NFTT
b) cAMP- and cGMP-dependent protein kinase phosphorylation site: 306 to 309 RRQS
c) Protein kinase C phosphorylation site: 95 to 97 SLR; 305 to 307 TRR; 366 to 368 SIR; 376 to 378 SSR; 390 to 392 SLK.
d) Casein kinase II phosphorylation site: 275 to 278 SDVE; 309 to 312 SNYE; 316 to 319 SQAE; 344 to 347 SEPE; 377 to 380 SRAE; 390 to 393 SLKD; 394 to 397 TYLE.
e) Amidation site: 386 to 389 VGRR
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: 1-52
b) 7 tentative TMs, membrane topology: type 3a
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
(1) ModBase predicted 3D structure of Q8BGF8 from UCSC Genome Sorter: none.
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,917Da, pI=6.99 (NP_083805).
FUNCTION
Ontology
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
(1) Biological process: carbohydrate transport and metabolism.
(2) Nucleotide-sugar transporter.
(3) Posttranslational modification.
(4) Platelet dense granule biogenesis.
Location
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
Endoplasmic reticulum membrane.
Interaction
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
A component of UDP-glucuronate transporter homohexamer. View Reactome details .
Pathway
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
Slc35d3 may participate in Xenobiotic metabolism as shown in KEGG Pathways.
MUTATION
Allele or SNP
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
Slc35d3 is described in MGI:1923407 .
No SNPs deposited in dbSNP.
Distribution
Location Genomic cDNA Protein Type Strain Reference Exon 1 ins 30G (IAP with a GGCAGCT repeat) 21 bp substitution of exon 1 M1-T7 substitution substitution C3H/HeSnJ-ash Chintala, et al
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_029529.)
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 1 | ins 30G (IAP with a GGCAGCT repeat) | 21 bp substitution of exon 1 | M1-T7 substitution | substitution | C3H/HeSnJ-ash | Chintala, et al |
Effect
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes.
The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724
Insertion of an IAP element into exon 1 of the Slc35d3 gene
alters the 5' terminal sequence of the Slc35d3 cDNA to introduce a new IAP derived
ATG start site in the ash-Roswell mutant. This
results in substitution of 21 new in-frame coding nucleotides for the 30 coding nucleotides found in control C3H/HeSnJ DNA. The
predicted result is the substitution of 7 new N-terminal amino acids in mutant
Slc35d3.
Northern blot analyses of poly(A)-RNA from brain revealed lack of expression of
the normal 2.6 kb Slc35d3 mRNA in mutant brain. However, this tissue
and other mutant tissues such as spleen exhibited greatly amplified expression of
an abnormal 2.2 kb transcript with additional transcripts at 3.1 and 3.8
kb. 8-2000 fold enhanced (compared to
levels in corresponding control C3H tissues) expression of Slc35d3 in multiple
tissues were observed in ash-Roswell mutants. This indicates that stable, highly
expressed mutant Slc35d3 transcripts exist in multiple tissues in ash-Roswell mice.
The enhanced Slc35d3 expression does not cause gain of function
since Slc35d3+/- heterozygotes display normal platelet dense granule numbers and
serotonin levels. Rather, the mutation behaves as a typical recessive loss of
function genetic trait in platelets, implying that the mutant protein (assuming it is
stable) is nonfunctional (Chintala, et al).
PHENOTYPE
Mutation in the Slc35d3 gene is the cause of recessive roswell (ros) mutant (Chintala, et al). The ros allele arose spontaneously in the C3H/HeSnJ-ash strain. The strain is described in more detail in JAX Mice database (C3H/HeSnJ-Rab27a/J). An ashen mutant line originally obtained from The Jackson Laboratory and subsequently maintained at Roswell Park Cancer Institute (thereafter referred to as ash-Roswell) displays the typical features of HPS including a prolonged bleeding time accompanied by substantial platelet dense granule deficiency in whole mount electron microscopy. However, the original ashen mutant line independently maintained at The Jackson Laboratory has normal bleeding times and normal concentrations of platelet dense granule components. A mutation in a second gene, Slc35d3, which encodes an orphan transporter with significant sequence homology to sugar nucleotide transporters, has occurred in the ash-Roswell mutant line and is the cause of its platelet dysfunction. Slc35d3 causes platelet dysfunction by regulating the contents of platelet dense granules. It differs from well-established HPS and CHS genes in that its effects on lysosome-related organelles are specific to platelet dense granules with no effect on pigmentation (melanosomes) or lysosomes. The ash-Roswell mouse mutant is an appropriate model for human congenital isolated delta-storage pool deficiency.(Chintala, et al).
REFERENCE
- Chintala S, Tan J, Gautam R, Rusiniak ME, Guo X, Li W, Gahl WA, Huizing M, Spritz RA, Hutton S, Novak EK, Swank RT. The Slc35d3 gene, encoding an orphan nucleotide sugar transporter, regulates platelet dense granules. Blood. 2006 Oct 24; [Epub ahead of print] PMID: 17062724