GENOMIC
Mapping
XqF3. View the map and BAC clones (data from UCSC genome browser).
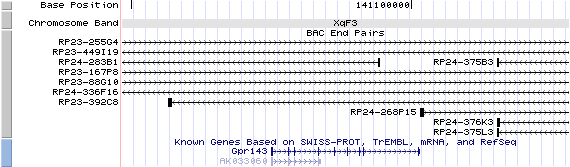
Structure
(assembly 10/03)
Gpr143/NM_010951: 9 exons, 26,661bp, ChrX: 141,074,902 - 141,101,562.
The figure below shows the structure of the Gpr143 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
Tissue-specific control of Oa1 transcription lies within a region of 617 bp that contains the E-box bound by Mitf (Vetrini, et al).
TRANSCRIPT
RefSeq/ORF
Gpr143 (NM_010951), 1,584bp, view ORF and the alignment to genomic.
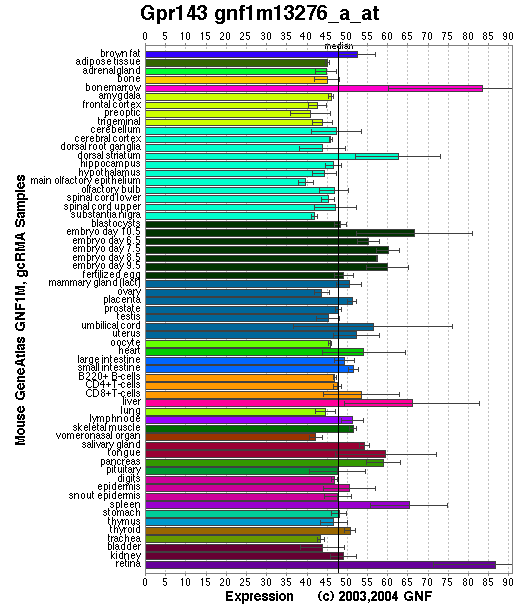
Expression Pattern
Tissue specificity: Pigment cells. A transcript of approximately 1.8 kb was readily detected. Oa1 expression can be detected at early stages (E10.5) of retinal pigment epithelium (RPE) development (Surace, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
G protein-coupled receptor 143 (NP_035081): 405aa, ExPaSy NiceProt view of Swiss-Prot:P70259.
Synonyms: Ocular albinism type 1 protein homolog; Moa1.
Ortholog
| Species | Mouse | Rat | Zebrafish | Mosquito | Fruitfly |
| GeneView | OA1 | LOC302619 | 393795 | 1277742 | CG4521/mthl1 |
| Protein | NP_000264 (424aa) | XP_228848 (464aa) | NP_957116 (412aa) | XP_317232 (321aa) | NP_573140 (676aa) |
| Identities | 77% /315aa | 94% /347aa | 54% /219aa | 28% /92aa | 20% /45aa |
View multiple sequence alignment (PDF files) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) low complexity 34 - 56
b) transmembrane 123 - 145
c) transmembrane 154 - 176
d) transmembrane 191 - 213
e) transmembrane 246 - 268
f) transmembrane 288 - 310
(2) Transmembrane domains predicted by SOSUI: eight transmembrane helices detected.
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 3 | SPRLGIFCCPTWDAATQLVLSFQ | 25 | SECONDARY | 23 |
| 2 | 30 | HALCLGSGTLRLVLGLLQLLSG | 51 | SECONDARY | 22 |
| 3 | 68 | VHILRAATACDLLGCLGIVIRST | 90 | SECONDARY | 23 |
| 4 | 118 | GSAMWIQLLYSACFWWLFCYAVD | 140 | PRIMARY | 23 |
| 5 | 156 | LYHIMAWGLAVLLCVEGAVSLYY | 178 | PRIMARY | 23 |
| 6 | 194 | HYVTTYLPLLLVLVANPILLHKT | 216 | SECONDARY | 23 |
| 7 | 248 | KIMLVLIACWLSNIINESLLFYL | 270 | PRIMARY | 23 |
| 8 | 291 | TWFIMGILNPAQGLLLSLAFYG | 312 | PRIMARY | 22 |
Motif/Site
(1) Predicted results by ScanProsite:
a) Protein kinase C phosphorylation site : [occurs frequently]
3 - 5: SpR,
38 - 40: TlR,
50 - 52: SgR,
280 - 282: SlK.
b) Amidation site : [occurs frequently]
50 - 53: sGRR,
225 - 228: kGRK.
c) Casein kinase II phosphorylation site : [occurs frequently]
75 - 78: TacD,
182 - 185: SrcE,
232 - 235: TenE,
335 - 338: SaaE,
364 - 367: TsdE,
379 - 382: StvE.
d) N-glycosylation site : [occurs frequently]
101 - 104: NISN,
106 - 109: NATD,
263 - 266: NESL.
e) N-myristoylation site : [occurs frequently]
187 - 192: GLdhAI,
303 - 308: GLllSL,
361 - 366: GGhtSD.
(2) Predicted results of subprograms by PSORT II:
a) Seems to have a cleavable signal peptide: 1 - 51.
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: found XXRR-like motif in the N-terminus: ASPR
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1)No ModBase entry found.
(2) 3D models predicted by SPARKS (fold recognition) below.View the models by PDB2MGIF.
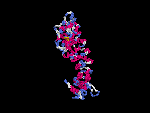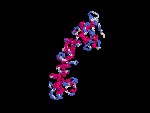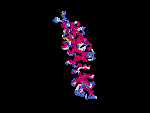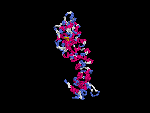
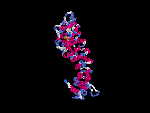
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=44,471Da, pI=8.15 (NP_035081).
FUNCTION
Ontology
a) Biological process: eye pigment biosynthesis; visual perception
b) Biological process: G-protein coupled receptor protein signaling pathway
c) Integral to membrane
d) Belongs to family OA of G-protein coupled receptors.
Location
Late endo-lysosomal compartments (Shen, et al ).
Melanosomal membrane (Schiaffino, et al (1996; 1999)).
Interaction
OA1 protein is a conserved integral membrane protein with seven transmembrane domains that has weak similarities to G protein-coupled receptors (GPCRs), which participate in the most common signal transduction system at the plasma membrane. It binds heterotrimeric G proteins, which suggests that OA1 GPCR-mediated signal transduction systems also operate at the internal membranes in mammalian cells (Schiaffino, et al (1999) ). OA1 colocalizes and coprecipitates with arrestins, which downregulate the signaling of OA1 by specifically reducing its expression levels (Innamorati, et al). Oa1 is a target of MITF, and tissue-specific control of Oa1 transcription lies within a region of 617 bp that contains the E-box bound by Mitf (Vetrini, et al).
The Drosophila homolog, CG4521, has no entries in the CuraGen Drosophila interaction database.
Pathway
The OA1 gene product is a membrane glycoprotein that may play a role in controlling melanosome growth and maturation. OA1 protein function in the late stage of melanosome development.
MUTATION
Allele or SNP
1 phenotypic alleles described in MGI:107193. No spontaneous mutation has been found.
Distribution
A PGK-hprt expression cassette replaced exon 1 including the start codon of the Oa1 gene in the Gpr143tm1Inc allele (Incerti, et al).
Effect
Northern analysis of primary melanocyte cultures did not detect Oa1 transcript in homozygous mutant neonates (Incerti, et al).
PHENOTYPE
Incerti, et al generated the Oa1-deficient mice by gene targeting (KO). The KO males are viable, fertile and phenotypically indistinguishable from the wild-type littermates. Ophthalmologic examination shows hypopigmentation of the ocular fundus in mutant animals compared with wild-type. Analysis of the retinofugal pathway reveals a reduction in the size of the uncrossed pathway, demonstrating a misrouting of the optic fibres at the chiasm, as observed in OA1 patients (OMIM 300500). Microscopic examination of the RPE shows the presence of giant melanosomes comparable with those described in OA1 patients. Ultrastructural analysis of the RPE cells, suggests that the giant melanosomes may form by abnormal growth of single melanosomes, rather than the fusion of several, shedding light on the pathogenesis of ocular albinism . For more information about this oa1-KO phenotype, view the Mouse Locus Catalog #Gpr143.
REFERENCE
- Incerti B, Cortese K, Pizzigoni A, Surace EM, Varani S, Coppola M, Jeffery G, Seeliger M, Jaissle G, Bennett DC, Marigo V, Schiaffino MV, Tacchetti C, Ballabio A. Oa1 knock-out: new insights on the pathogenesis of ocular albinism type 1. Hum Mol Genet 2000; 9: 2781-8. PMID: 11092754
- Innamorati G, Piccirillo R, Bagnato P, Palmisano I, Schiaffino MV. The melanosomal/lysosomal protein OA1 has properties of a G protein-coupled receptor. Pigment Cell Res 2006; 19: 125-35. PMID: 16524428
- Schiaffino MV, Baschirotto C, Pellegrini G, Montalti S, Tacchetti C, De Luca M, Ballabio A. The ocular albinism type 1 gene product is a membrane glycoprotein localized to melanosomes. Proc Natl Acad Sci U S A 1996; 93: 9055-60. PMID: 8799153
- Schiaffino MV, d'Addio M, Alloni A, Baschirotto C, Valetti C, Cortese K, Puri C, Bassi MT, Colla C, De Luca M, Tacchetti C, Ballabio A. Ocular albinism: evidence for a defect in an intracellular signal transduction system. Nat Genet 1999; 23: 108-12. PMID: 10471510
- Shen B, Rosenberg B, Orlow SJ. Intracellular distribution and late endosomal effects of the ocular albinism type 1 gene product: consequences of disease-causing mutations and implications for melanosome biogenesis. Traffic 2001; 2: 202-11. PMID: 11260525
- Surace EM, Angeletti B, Ballabio A, Marigo V. Expression pattern of the ocular albinism type 1 (Oa1) gene in the murine retinal pigment epithelium. Invest Ophthalmol Vis Sci 2000; 41: 4333-7. PMID: 11095635
- Vetrini F, Auricchio A, Du J, Angeletti B, Fisher DE, Ballabio A, Marigo V. The microphthalmia transcription factor (Mitf) controls expression of the ocular albinism type 1 gene: link between melanin synthesis and melanosome biogenesis. Mol Cell Biol 2004; 24: 6550-9. PMID: 15254223
- Vetrini F, Auricchio A, Du J, Angeletti B, Fisher DE, Ballabio A, Marigo V. The microphthalmia transcription factor (Mitf) controls expression of the ocular albinism type 1 gene: link between melanin synthesis and melanosome biogenesis. Mol Cell Biol 2004; 24: 6550-9. PMID: 15254223
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne: 07/29/2004
Updated by Wei Li: 04/06/2006