GENOMIC
Mapping
6qD3. View the map and BAC clones (data from UCSC genome browser).
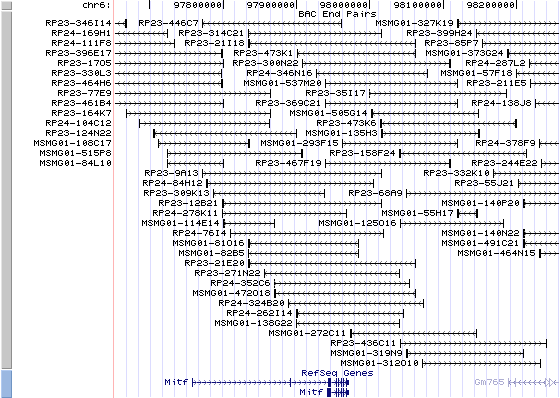
Structure
(assembly 02/2006)
Mitf (NM_008601): 9 exons, 26,911 bp, chr6:97957255-97984165.
The figure below shows the structure of the Mitf gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Mitf, seq2=human MITF) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Mitf (NM_008601), 1,921 bp, view ORF and the alignment to genomic.
Expression Pattern
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
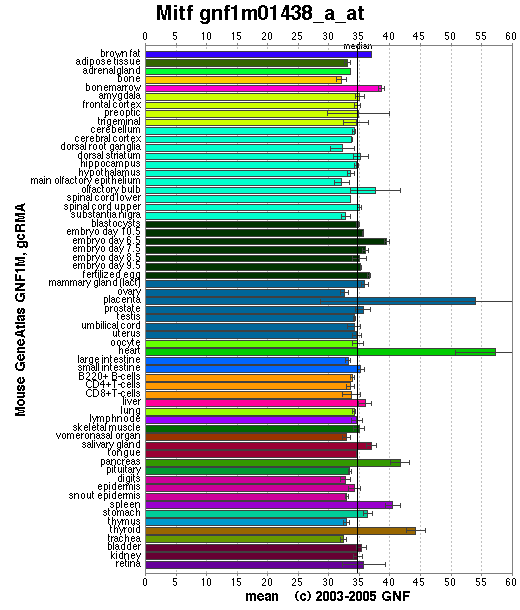
Browse more information in Entrez Gene, UCSC Gene Sorter, MGI.
PROTEIN
Sequence
microphthalmia-associated transcription factor (NP_032627): 419 aa, UniProtKB/Swiss-Prot entry Q32MU6.
Ortholog
Species Human
Chimpanzee Dog Rat Chicken GeneView
MITF
MITF
MITF
Mitf
CMI9
Protein
NP_937802 (520 aa)
XP_001138876 (520 aa)
XP_855594 (419 aa)
XP_001065702 (520 aa)
NP_990360 (468 aa)
Identities
378/409 (92%) 376/409 (91%) 390/419 (93%) 398/409 (97%) 355/409 (86%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) HLH: 210-263
b) low complexity: 334-351
c) low complexity: 400-415
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 323 to 326 NCSQ.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 406 to 409 RRSS.
c) Protein kinase C phosphorylation site:
Site : 404 to 406 SSR.
Site : 405 to 407 SRR.
d) Casein kinase II phosphorylation site:
Site : 14 to 17 THLE.
Site : 82 to 85 SNCE.
Site : 96 to 99 SRAE.
Site : 129 to 132 SYNE.
Site : 192 to 195 TESE.
Site : 339 to 342 TTLD.
Site : 356 to 359 TMPE.
Site : 372 to 375 SNLE.
Site : 411 to 414 SAEE.
e) N-myristoylation site:
Site : 167 to 172 GLTISN.
Site : 399 to 404 GASKTS.
f) Leucine zipper pattern:
Site : 267 to 288 LENRQKKLEHANRHLLLRVQEL.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1)
ModBase predicted 3D structure of Q32MU6 from UCSC Genome Sorter:
 From left to right: Front, Top, and Side views of predicted protein.
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=46,768.8Da, pI=5.84.
| Species | Human | Chimpanzee | Dog | Rat | Chicken |
| GeneView | MITF | MITF | MITF | Mitf | CMI9 |
| Protein | NP_937802 (520 aa) | XP_001138876 (520 aa) | XP_855594 (419 aa) | XP_001065702 (520 aa) | NP_990360 (468 aa) |
| Identities | 378/409 (92%) | 376/409 (91%) | 390/419 (93%) | 398/409 (97%) | 355/409 (86%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) HLH: 210-263
b) low complexity: 334-351
c) low complexity: 400-415
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 323 to 326 NCSQ.
b) cAMP- and cGMP-dependent protein kinase phosphorylation site:
Site : 406 to 409 RRSS.
c) Protein kinase C phosphorylation site:
Site : 404 to 406 SSR.
Site : 405 to 407 SRR.
d) Casein kinase II phosphorylation site:
Site : 14 to 17 THLE.
Site : 82 to 85 SNCE.
Site : 96 to 99 SRAE.
Site : 129 to 132 SYNE.
Site : 192 to 195 TESE.
Site : 339 to 342 TTLD.
Site : 356 to 359 TMPE.
Site : 372 to 375 SNLE.
Site : 411 to 414 SAEE.
e) N-myristoylation site:
Site : 167 to 172 GLTISN.
Site : 399 to 404 GASKTS.
f) Leucine zipper pattern:
Site : 267 to 288 LENRQKKLEHANRHLLLRVQEL.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q32MU6 from UCSC Genome Sorter:


 From left to right: Front, Top, and Side views of predicted protein.
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=46,768.8Da, pI=5.84.
FUNCTION
Ontology
(1) Biological process: DNA binding, DNA-dependent regulation of transcription, sensory perception of sound.
(2) Transcription factor activity.
(3) Sequence-specific (E-boxes) (5'-CACGTG-3') DNA binding.
(4) Plays a critical role in the differentiation of various cell types as neural crest-derived melanocytes, mast cells, osteoclasts and optic cup-derived retinal pigment epithelium.
Location
Nucleus.
Interaction
Transcription factor for tyrosinase and tyrosinase-related protein 1. Efficient DNA binding requires dimerization with another bHLH protein. Binds DNA in the form of homodimer or heterodimer with either TFE3, TFEB or TFEC. Phosphorylation at Ser-405 significantly enhances the ability to bind the tyrosinase promoter.
Interactions in HPRD.
View co-occured partners in literature searched by PPI Finder.
Pathway
MITF is involved in Wnt receptor signaling pathway. MITF regulates tyrosinase and c-KIT gene. Melanogenesis pathway and Melanoma pathway in KEGG. BioCarta from NCI Cancer Genome Anatomy Project - Melanocyte Development and Pigmentation Pathway. During melanocyte development, the Mitf connects three major signaling pathways involving WNT, KIT, and EDNRB. More details about these signalingpathways are reviewed by Hou et al.
MUTATION
Allele or SNP
29 phenotypic alleles of Mitf are described in MGI:104554.
SNPs deposited in dbSNP Build 128.
Distribution
The 29 phenotypic alleles inclede knock-out and others (4), knock-in (2), spontaneous (13), chemically induced (7), radiation induced (3).
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_008601. view ORF here.)
Effect
Mutations at this locus affect development of melanocytes, mast cells, osteoclasts and pigmented epithelium. Mutants variably display lack of pigment in coat and eye, microphthalmia, hearing loss, bone resorption anomalies, mast cell deficiency and lethality.
PHENOTYPE
Mutations at the Mitf locus affect eye size, pigmentation, and the capacity for secondary bone resorption. Mice homozygous for the white allele (MitfMi-wh) display an overall absence of pigment cells with the exception of the retina which expresses a few giving the eye a small amount of pigment. Homozygotes show slight microphthalmia but a normal skeleton. Heterozygotes have a diluted coat color, light ears, a white belly spot, and in rare cases a dorsal spot. In addition, they display abnormalitites of both the cochlear and vestibular portions of the inner ear. The Mi-wh allele arose spontaneously in the C57BL/6J strain. The strain is described in more detail in JAX Mice database (C57BL/6J-MitfMi-wh/J). Some of these alleles are being used to model the human diseases Waardenburg syndrome IIa and Tietz syndrome. Mitf is expressed in developing neural crest-derived melanoblasts and in the neuroepithelium-derived retinal pigmented epithelium (RPE) of the eye beginning at E10, and Mitf mutations cause loss of neural crest-derived melanoblasts and abnormal eye development.
REFERENCE
- Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res 2008; 18:1163-76. Review. PMID: 19002157