GENOMIC
Mapping
13qA1; View the map and BAC clones (data from UCSC genome browser).
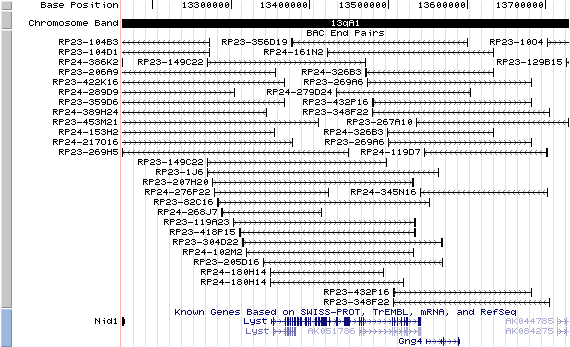
Structure
(assembly 10/03)
Lyst/NM_019788: 52 exons, 189,325bp, Chr13: 13,350,721 - 13,540,045.
The figure below shows the structure of the lyst gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (human and mouse) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Lyst (NM_010748), 11,817bp, view ORF and the alignment to genomic.
Expression Pattern
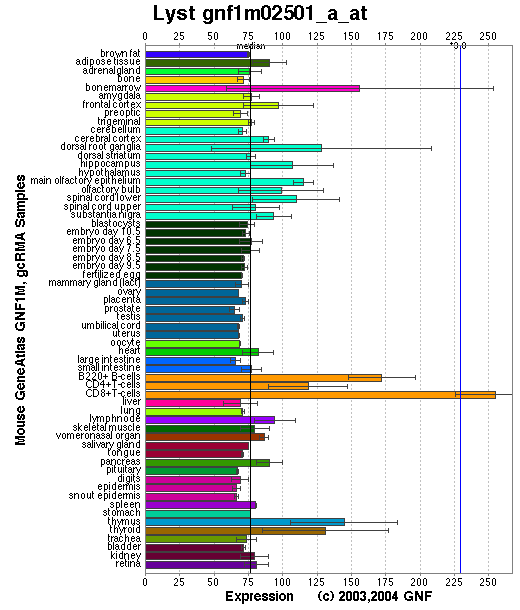
Tissue specificity: Varied expression, highest and lowest found in thymus and small intestine respectively. In mRNA from mouse spleen, the 12-14kb transcript is barely detectable, but smaller transcript of 7kb is abundant (Barbosa, et al).
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
PROTEIN
Sequence
Lysosomal trafficking regulator (NP_034878): 3788aa, ExPaSy NiceProt view of Swiss-Prot:P97412.
Synonyms: beige protein; CHS1 homolog.
Ortholog
| Species | Human | Rat | Zebrafish | HoneyBee | Fruitfly |
| GeneView | CHS1 | Lyst | 06386 | LOC408774 | CG11814 |
| Protein | NP_000072 (3801aa) | NP_445970 (3788aa) | 25336 (1292aa) | XP_392306 (3219aa) | NP_647681 (2517aa) |
| Identities | 84%/3228aa | 93%/3538aa | 49%/655aa | 33%/501aa | 25%/588aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) low complexity 26 - 36
b) low complexity 72 - 82
c) low complexity 399 - 412
d) low complexity 1333 - 1344
e) low complexity 2296 - 2308
f) low complexity 2428 - 2446
g) low complexity 2535 - 2547
h) Pfam:Beach: 3119 - 3409
i) Pfam:WD40: 3540 - 3580, 3592 - 3631, 3634 - 3677, 3725 - 3766
(2) Transmembrane domains predicted by SOSUI: None.
(3) Graphical view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a) BEACH domain profile :
3107 - 3409: score=137.477:
b) Trp-Asp (WD) repeats profile :
3599 - 3633: score=9.840:
c) Trp-Asp (WD) repeats circular profile :
3599 - 3775: score=15.478:
d) Trp-Asp (WD) repeats signature :
3618 - 3632: MISVsrDgtCiVWDL
e) cAMP- and cGMP-dependent protein kinase phosphorylation site : [occurs frequently]
147 - 150: RKsT,
884 - 887: KRkT,
1627 - 1630: RKtS,
2575 - 2578: KRrS,
2611 - 2614: RRmS.
f) Amidation site : [occurs frequently]
1008 - 1011: qGRR,
1426 - 1429: rGKK,
1487 - 1490: rGKR,
1598 - 1601: qGKK.
g) Tyrosine kinase phosphorylation site : [occurs frequently]
1206 - 1214: KllgEeegY,
3092 - 3098: Kvr.Ddv.Y,
3142 - 3150: RsfnDlmqY,
3185 - 3191: Kek.Edr.Y,
3190 - 3197: Ryv.DtykY,
3196 - 3202: Kyl.Eee.Y.
h) Leucine zipper pattern : [occurs frequently]
2361 - 2382: LknrgfsLlanqlyLhrgtqeL
i) Bipartite nuclear targeting sequence : [occurs frequently]
144 - 160:
RRqrksthrysvrdark,
484 - 500:
KKvkseqlhhsmctrkr,
871 - 887:
RKfyaslrepdpkkrkt.
j) Tyrosine sulfation site : [occurs frequently]
536 - 550:
taegdvqYperccci,
921 - 935:
esedtsgYdsppsep,
1207 - 1221:
llgeeegYeadsesn,
3049 - 3063:
pasfswtYeeikevh.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: found KKXX-like motif in the C-terminus: PAKR
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: none
i) Dileucine motif in the tail: none
3D Model
(1) ModBase entry found, results here.
(2)ModBase Predicted Comparative 3D Structure of P97412 from UCSC Genome Sorter.
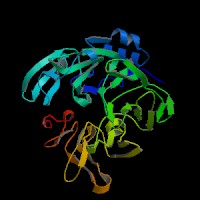
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=425,287Da, pI=6.15 (NP_034878).
FUNCTION
Ontology
a) Biological process: intracellular protein transport from endosome to lysosome transport
b) Involved in cellular defense response
c) Component of endosome
d) Associated with microtubule cytoskeleton.
Location
Cytoplasmic.
Interaction
Twenty-one LYST-interacting proteins (LIPs) were identified in yeast two-hybrid screens by using fourteen cDNA fragments, covering the entire coding domain of LYST, as baits to screen five human cDNA libraries. Four confirmed interactions were with proteins important in vesicular transport and signal transduction (the SNARE-complex protein HRS, 14-3-3 tau, casein kinase II beta subunit, and troponin I) (Tchernev, et al). Fujita, et al identified the SBP1 by yeast two-hybrid screening, with the N-terminal half (1-157) of human SBP1 identical to lyst-interacting protein 5. SBP1 binds directly to SKD1, a member of the AAA-ATPase family and one of the mammalian class E Vps (vacuolar protein sorting) proteins, through its C-terminal region (198-309). They proposed that the SKD1-SBP1 complex, together with lyst protein, may function in endosomal membrane transport.
Lyst drosophila homolog CG11814 interaction information shown in CuraGen interaction database.
Pathway
The beige protein may be required for sorting endosomal resident proteins into late multivesicular endosomes by a mechanism involving microtubules.
MUTATION
Allele or SNP
9 phenotypic alleles described in MGI:107448.
Distribution
| Location | Genomic | cDNA | Protein | Type | Strain | Reference |
| Exon 4 | del ~5kb | ?~3715G del partial exon 4 and exons 5~6 | del ~400aa | gross deletion | bg-11J (B6) | Barbosa, et al |
| Exon 22 | 6832C>T | 6832C>T | R2278X | nonsense | bg-8J (DBA/2J) | Perou, et al (1996) |
| Intron 22 | 7029Ains L1-bg (1097bp 3'-LINE1, 45A, 11-14bp dup) | 7029Ains 116bp or 325 bp | K2343ins 116bp or 325 bp | frame-shift | bg (B6) | Perou, et al (1997) |
| Intron 25 | splicing donor, +2T>C | 7196Adel 231 bp | E2399del 77aa (exon 25) | in-frame | bg-grey (ENU) | Runkel, et al |
(Numbering of genomic and cDNA sequence is based on the start codon of RefSeq NM_010748.)
Effect
All the identified mutations are predicted to produce truncated proteins. Western blot analysis suggests that the Lyst(bg-grey) mutation causes instability of the LYST protein (Runkel, et al).
PHENOTYPE
Defects in Lyst are the cause of beige (bg) (Barbosa, et al; Perou, et al (1996; 1997)), an autosomal recessive disorder characterized by hypopigmentation, bleeding, immune cell dysfunction resulting in increased susceptibility to bacterial, viral, and parasitic infections and decreased cytotoxic activity against tumor cells, abnormal intracellular transport to and from the lysosome (missorting of proteins, such as elastase, glucuronidase and cathepsin G), and giant inclusion bodies in a variety of cell types. The mice with the bg mutation (Brandt, et al) exhibit abnormalities similar to human Chediak-Higashi syndrome (OMIM 214500). However, the bg mice do not develop signs of an accelerated phase. Life span is severely reduced in the beige (McGarry, et al). Giant lamellar body degeneration (GLBD) was found in both ep and bg mice soon after birth, and increased in severity as the mice grew older. Aged ep mice with less severe GLBD than bg mice of comparable ages also had a slight tendency to develop interstitial inflammation but no fibrosis. Ep and bg mice, especially the latter, may be useful mouse models of HPS-associated interstitial pneumonia (HPSIP) (Tang, et al). For more details of the beige phenotype, view the Mouse Locus Catalog #Lyst. The beige mutation arose from C3H/Rl (radiation) and was introduced to the C57BL/6J strain. The strain is described in more detail in JAX Mice database (B6.Cg-Lystbg/J).
Two spontaneous mutant beige rats, with phenotypes resembling human Chediak-Higashi syndrome (CHS), were found independently in two inbred strains. Both beige mutations were identified to be recessive alleles in the Lyst locus on rat chromosome 17 and the alleles were denoted Lyst(bg) and Lyst(bg-Kyo). An allele-specific genotyping method was developed to discriminate these alleles (Masui, et al). Chediak-Higashi syndrome in Japanese black cattle (Wagyu) is a hereditary disease with prolonged bleeding time and partial albinism. The CHS locus was mapped on proximal region of bovine chromosome 28. The H2015R mutation of the bovine LYST homolog is likely the cause of CHS in Wagyu cattle (Kunieda, et al). Loss of LvsB, the Dictyostelium protein most similar to LYST/Beige, resulted in the formation of enlarged vesicles that by multiple criteria appeared to be acidic lysosomes, suggesting that LvsB acts as a negative regulator of fusion and supporting the notion that LvsB/LYST/Beige function in a similar manner to regulate lysosome biogenesis (Harris, et al). In addition, many other mammalian species, including mink, cat, fox, and killer whale, have been described similar symptoms to human CHS.
REFERENCE
- Barbosa MD, Nguyen QA, Tchernev VT, Ashley JA, Detter JC, Blaydes SM, Brandt SJ, Chotai D, Hodgman C, Solari RC, Lovett M, Kingsmore SF. Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature 1996; 382: 262-5. PMID: 8717042
- Brandt EJ, Swank RT. The Chediak-Higashi (beige) mutation in two mouse strains. Allelism and similarity in lysosomal dysfunction. Am J Pathol 1976; 82: 573-88. PMID: 1258977
- Fujita H, Umezuki Y, Imamura K, Ishikawa D, Uchimura S, Nara A, Yoshimori T, Hayashizaki Y, Kawai J, Ishidoh K, Tanaka Y, Himeno M. Mammalian class E Vps proteins, SBP1 and mVps2/CHMP2A, interact with and regulate the function of an AAA-ATPase SKD1/Vps4B. J Cell Sci 2004; 117: 2997-3009. PMID: 15173323
- Harris E, Wang N, Wu Wl WL, Weatherford A, De Lozanne A, Cardelli J. Dictyostelium LvsB mutants model the lysosomal defects associated with Chediak-Higashi syndrome. Mol Biol Cell 2002; 13: 656-69. PMID: 11854420
- Kunieda T, Nakagiri M, Takami M, Ide H, Ogawa H. Cloning of bovine LYST gene and identification of a missense mutation associated with Chediak-Higashi syndrome of cattle. Mamm Genome 1999; 10: 1146-9. PMID: 10594238
- Masui N, Mori M, Nishikawa T, Takagi Y, Asai H, Yanabe M, Yamasaki K, Serikawa T, Sato K. An allele-specific genotyping method for rat lyst (lysosomal trafficking regulator) gene. Exp Anim 2004; 53: 77-80. PMID: 14993748
- McGarry MP, Reddington M, Novak EK, Swank RT. Survival and lung pathology of mouse models of Hermansky-Pudlak syndrome and Chediak-Higashi syndrome. Proc Soc Exp Biol Med. 1999; 220: 162-8. PMID: 10193444
- Perou CM, Moore KJ, Nagle DL, Misumi DJ, Woolf EA, McGrail SH, Holmgren L, Brody TH, Dussault BJ Jr, Monroe CA, Duyk GM, Pryor RJ, Li L, Justice MJ, Kaplan J. Identification of the murine beige gene by YAC complementation and positional cloning. Nat Genet 1996; 13: 303-8. PMID: 8673129
- Perou CM, Pryor RJ, Naas TP, Kaplan J. The bg allele mutation is due to a LINE1 element retrotransposition. Genomics 1997; 42: 366-8. PMID: 9192864
- Runkel F, Bussow H, Seburn KL, Cox GA, Ward DM, Kaplan J, Franz T. Grey, a novel mutation in the murine Lyst gene, causes the beige phenotype by skipping of exon 25. Mamm Genome. 2006; 17: 203-10. PMID: 16518687
- Tang X, Yamanaka S, Miyagi Y, Nagashima Y, Nakatani Y. Lung pathology of pale ear mouse (model of Hermansky-Pudlak syndrome 1) and beige mouse (model of Chediak-Higashi syndrome): severity of giant lamellar body degeneration of type II pneumocytes correlates with interstitial inflammation. Pathol Int 2005; 55:137-43.PMID: 15743322
- Tchernev VT, Mansfield TA, Giot L, Kumar AM, Nandabalan K, Li Y, Mishra VS, Detter JC, Rothberg JM, Wallace MR, Southwick FS, Kingsmore SF. The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol Med 2002; 8: 56-64. PMID: 11984006
EDIT HISTORY:
Created by Wei Li & Jonathan Bourne: 07/13/2004
Updated by Wei Li: 06/25/2005
Updated by Wei Li: 04/06/2006