GENOMIC
Mapping
1qc4. View the map and BAC clones (data from UCSC genome browser).
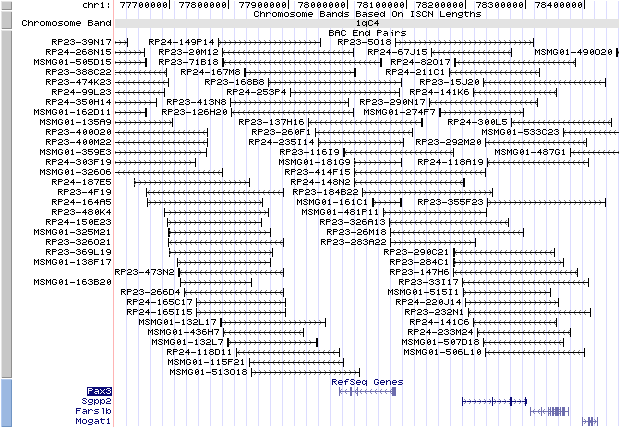
Structure
(assembly 02/2006)
Pax3 (NM_008781): 8 exons, 94,445 bp, chr1:77985839-78080283.
The figure below shows the structure of the Pax3 gene (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Pax3, seq2=human PAX3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Pax3 (NM_008781), 2,444 bp, view ORF and the alignment to genomic.
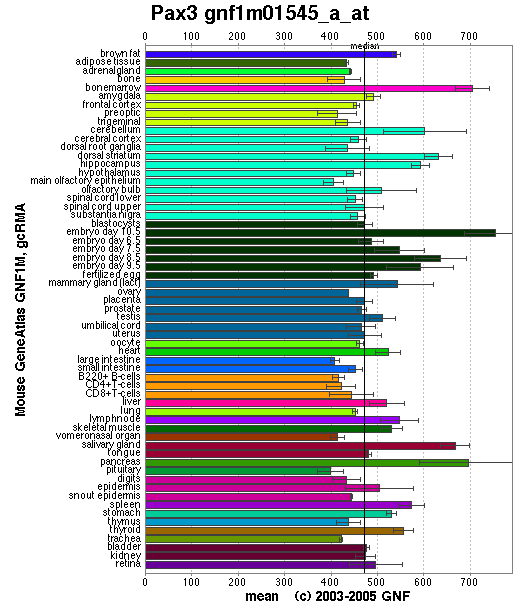
Expression Pattern
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
Browse more information in Entrez Gene, UCSC Gene Sorter, MGI.
PROTEIN
Sequence
paired box gene 3 (NP_032807): 479 aa, UniProtKB/Swiss-Prot entry P24610.
Ortholog
Species
Human Chimpanzee Dog Rat Chicken GeneView
PAX3
PAX3
PAX3
Pax3
PAX3
Protein
NP_852124 (505 aa)
XP_001165390 (505 aa)
XP_545664 (468 aa)
XP_343602 (476 aa)
NP_989600 (484 aa)
Identities
467/474 (98%) 467/474 (98%) 433/448 (96%) 469/481 (97%) 451/474 (95%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) PAX 34-159
b) low complexity 164-185
c) HOX 219-281
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)Protein kinase C phosphorylation site:
Site : 95 to 97 SIR.
Site : 180 to 182 SEK.
Site : 193 to 195 SER.
Site : 268 to 270 SNR.
Site : 358 to 360 STR.
Site : 430 to 432 SQR.
b) Casein kinase II phosphorylation site:
Site : 110 to 113 TTPD.
Site : 205 to 208 SDID.
Site : 209 to 212 SEPD.
Site : 248 to 251 TREE.
Site : 346 to 349 SNAD.
Site : 365 to 368 SYTD.
c) Tyrosine kinase phosphorylation site:
Site : 236 to 243 RAFERTHY.
d) N-myristoylation site:
Site : 43 to 48 GVFING.
Site : 66 to 71 GIRPCV.
Site : 99 to 104 GAIGGS.
Site : 394 to 399 GLLTNH.
Site : 418 to 423 GLEPTT.
e) 'Homeobox' domain signature:
Site : 252 to 275 LAQRAKLTEARVQVWFSNRRARWR.
f) Paired domain signature.:
Site : 68 to 84 RPCVISRQLRVSHGCVS.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KKXX-like motif in the C-terminus: KPWT
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1)
ModBase predicted 3D structure of P24610 from UCSC Gene Sorter:
From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=52,890.4Da, pI=9.01.
| Species | Human | Chimpanzee | Dog | Rat | Chicken |
| GeneView | PAX3 | PAX3 | PAX3 | Pax3 | PAX3 |
| Protein | NP_852124 (505 aa) | XP_001165390 (505 aa) | XP_545664 (468 aa) | XP_343602 (476 aa) | NP_989600 (484 aa) |
| Identities | 467/474 (98%) | 467/474 (98%) | 433/448 (96%) | 469/481 (97%) | 451/474 (95%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) PAX 34-159
b) low complexity 164-185
c) HOX 219-281
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)Protein kinase C phosphorylation site:
Site : 95 to 97 SIR.
Site : 180 to 182 SEK.
Site : 193 to 195 SER.
Site : 268 to 270 SNR.
Site : 358 to 360 STR.
Site : 430 to 432 SQR.
b) Casein kinase II phosphorylation site:
Site : 110 to 113 TTPD.
Site : 205 to 208 SDID.
Site : 209 to 212 SEPD.
Site : 248 to 251 TREE.
Site : 346 to 349 SNAD.
Site : 365 to 368 SYTD.
c) Tyrosine kinase phosphorylation site:
Site : 236 to 243 RAFERTHY.
d) N-myristoylation site:
Site : 43 to 48 GVFING.
Site : 66 to 71 GIRPCV.
Site : 99 to 104 GAIGGS.
Site : 394 to 399 GLLTNH.
Site : 418 to 423 GLEPTT.
e) 'Homeobox' domain signature:
Site : 252 to 275 LAQRAKLTEARVQVWFSNRRARWR.
f) Paired domain signature.:
Site : 68 to 84 RPCVISRQLRVSHGCVS.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: KKXX-like motif in the C-terminus: KPWT
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of P24610 from UCSC Gene Sorter:



From left to right: Front, Top, and Side views of predicted protein.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=52,890.4Da, pI=9.01.
FUNCTION
Ontology
(1) Biological process: DNA-dependent regulation of transcription, neural crest cell migration, neural tube closure, sensory perception of sound.
(2) Transcription factor activity.
(3) Sequence-specific DNA binding.
(4) Embryogenesis and oncogenesis.
Location
Nucleus.
Interaction
It can bind to DNA as a heterodimer with PAX7. Interacts with DAXX.
View interacting proteins in IntAct .
View co-occured partners in literature searched by PPI Finder.
Pathway
PAX3 is involved in RNA polymerase II transcription factor activity. PAX3 synergistically transactivates the promoter of Mitf with SOX10. Mitf protein in turn regulates the tyrosinase and c-KIT gene.
MUTATION
Allele or SNP
26 phenotypic alleles of Pax3 are described in MGI:97487 .
SNPs deposited in dbSNP Build 128.
Distribution
The 26 phenotypic alleles inclede knock-out (1), knock-in and others (13), spontaneous (5), chemically induced (2), radiation induced (6).
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_008781. view ORF here.)
Effect
Effects on homzygotes for mutations in this gene vary in severity and include embryonic to perinatal death, malformations of neural tube, spinal ganglia, heart, vertebral column, hindbrain and limb musculature. Heterozygotes have white belly spots and variable spotting on the back and extremeties.
PHENOTYPE
Mutation in the Pax3 gene is the cause of Splotch mutant (Sp). The Sp allele arose spontaneously in the C57BL/6J strain. The strain is described in more detail in JAX Mice database (C57BL/6J-Pax3Sp/J). Mice homozygous for the Pax3Sp die at E13 due to neural tube defects and other malformations. Heterozygous Sp mice show white spotting on the belly and occasionally on the back, feet, and tail. The Splotch mutant is used as models for human neural tube defects (Greene et al) and Waardenburg syndrome types 1 and 3.
REFERENCE
- Greene ND, Massa V, Copp AJ. Understanding the causes and prevention of neural tube defects: Insights from the splotch mouse model. Birth Defects Res A Clin Mol Teratol 2009; 85:322-30. PMID: 19180568