CONREAL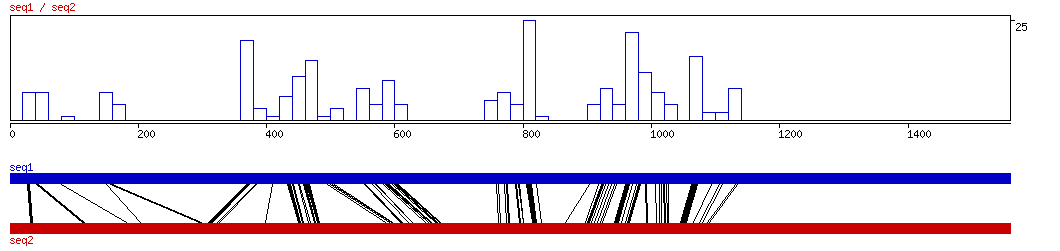
CONREAL ALIGNMENT 10 20 30 40
seq1 cttgagcaaattggtggatggtggtg-----TCATTTACTGaaacg------------------------------------------------------
| | ||||||| || |
seq2 tactgaacctcttataaccttgaccatcttgTCATTTAAAAaagagggtgtcaccctggtagccagtcttaaagcaaaatgaatcctcattttggaagtc
10 20 30 40 50 60 70 80 90 100
50 60 70 80 90 1
seq1 ------------GAAAATACAGGatgagcagattaaaggtgaacatgcaa-------------------------------TGAATGaaattttcaattt
|||| | || |||| | || | || || || | || ||
seq2 tgcactcaaggaAAAAAAAAAGCatgactaaatatccaacttcctagctcagtagtcgtctttcctgagtagaaaaggctcGGAGTGttttaatcccctt
110 120 130 140 150 160 170 180 190 200
00 110 120 130 140 150
seq1 tgtgtatgttggtctgagatgccttttggatacatgttgtatacatgcttaCATGAGt------------------------------------------
| || | |
seq2 ttc------------------------------------------------CACGTGgaagggctggggatgtcacccactgccagagtgcttgtctagt
210 220 230 240 250
160 170 180 190 200 210
seq1 -------------------------------------------AAGCAACTGGTATGaattgacggagagggaggaaggacagggcagaagatagagctg
|| |||||| | |
seq2 atgccacaaaggatgcttgagatgtcccggggacaacgcggtgAAACAACTGAGAGG-------------------------------------------
260 270 280 290 300
220 230 240 250 260 270 280 290 300 310
seq1 tgggagtcatcggttccttgacagaatttacagcgtggtaatggatagactctatgctatccatctatggaacaacacatctggagaagagagaagagct
seq2 ----------------------------------------------------------------------------------------------------
320 330 340 350 360 370 380 390 400
seq1 ggggaggctggactgtagccttgggagtggctattaaaataggagaaggggtggAGCGGAAGGGAGAGAGGAACTATGgagtgcttagcaagagaa----
| ||||||| | || || | ||| | | | | |
seq2 ------------------------------------------------------AAAGGAAGGGGGCAAGCAAAAACAgaggagtaccctacaatagatg
310 320 330 340 350
410 420 430 44
seq1 -------------------------------------------GAAAGTGTtttaaggatggggat---------------------------GATAAAC
| ||| || | |||| || |||
seq2 acggctagactagcccagggaaagtgcatagagaggaggaggaGTCTGTGCacgtagttttgggacagaagggatgaaaggaagaaaaaaggtGAGCAAC
360 370 380 390 400 410 420 430 440 450
0 450 460 470 480 490
seq1 TGCATTGAaaGCCACTGAGAGGTGGAGAAAAAATCAGGGcagagaagtaataaaa---------------------------------------------
||| ||| ||||| |||| ||||||| || ||| |||| || |
seq2 TGCTCTGAggATAACTGAAAGGTTGAGAAAAGATTGACGcacagaataaagagggttggctcagtgggagagccttggtatgcaagaaaccgagttccac
460 470 480 490 500 510 520 530 540 550
500 510 520 530 540 550 560
seq1 ------------------------------GGCATCAGCAACATGGAagtcactggtgactcgcgaaaggcgatcacctttggtgtgTAGGGCACAAAAg
||| | |||||||||| |||||||| ||
seq2 ccccacaagggggcagaggaggggaaacatGGCTTTGGCAACATGGAgagatg----------------------------------AAGGGCACAGAAa
560 570 580 590 600 610 620
570 580 590 600 610 620 630 640 650
seq1 CCTGGGGGGGTAAAgag---------AGAGTGGAAAGTAAGAATGTCGAGACAGTCGactattattgccattttatgaatgaagaaaccgaggctcaata
|| ||||| | || | | |||||||| | | ||||||| || || | | || | ||| | || |
seq2 TTTGACGGGGTCAGtcaaagatctagAGTGGGCAAAGTAAGGGTTTGGAGACAGGTGatcatcaccgacaaaccccaatacggagtaccaaagcagtgtt
630 640 650 660 670 680 690 700 710 720
660 670 680 690 700 710 720 730 740 750
seq1 tttgactgtctcattcaaggtgaaagccagtggggtcagatcctggacctagaatccatgcaatgatttgtgcacatcgcctattgaagagagagatcca
|| | | | | | |
seq2 gttatagggaggcgcagccacccaggagattcccgcagcg------------------------------------------------------------
730 740 750 760
760 770 780 790 800 810 820 830 840 850
seq1 attCTCTTCCAaaacGTAAAAGCCAACACTTc----CCTCAAAGGAGGCttcga-GTTAAATCATCCTTTCACGATGCcgcttccattttaaagaaaaaa
||||||| | ||||||||| | |||||| || | | ||||||||||| || || || | | || ||||
seq2 ---CTCTTCCGacctTCGAAAGCCAACCAACccttcAACTAAAGGATGCaggggcCTGAAATCATCCTTCCAAGAAGCagtcataaccttggagaaggcc
770 780 790 800 810 820 830 840 850 8
860 870 880 890 900 910 920 930
seq1 aaaaaaaaacgagaaggcctggtgcgggataggaatgagccgaggagctactGGCCCTctcggtcgg----------------GGGAAGACCCGCCCCTT
||||| || | | | |||| ||| ||| |
seq2 ggcttcc---------------------------------------------CGCCCTgtctgcagagcgggcgagcgcgctcGCGAAGGCCCCGCCCCT
60 870 880 890 900 910
940 950 960 970 980 990 1000 1010
seq1 TTCCGCAGCCCGAGCCcggcgcag----TCCACTGCGGCTGCGCGAGCCTGGCGCGCGCG------------------GGGCGGGAACgcggaaGGGGCT
||| || |||||| | |||||||||||||| ||| |||||||| |||| | || | | ||||
seq2 CGGCGCCGCGCGAGCCtcccctgagcgcCGCACTGCGGCTGCGCCGGCCGGGCGCGCGGAggggggcggggacgcggaAGGCGCGGCGgccgggGAGGCT
920 930 940 950 960 970 980 990 1000 1010
1020 1030 1040 1050 1060 1070 1080 1090 1100 1110
seq1 GGGGTTGCGGCGCGGCGGCgaggaccagaccgggggcggggccggtagtgGGAGTGCGGGGCGCGCggtgacagcgcgggGTTGGCGGCgtggGACCCAG
| || |||||| |||| || | || | | |||||||| | || | |||| | || ||
seq2 GCGGCGGCGGCGGCGCGGTgaagcgagagcc-------------------GACGCGCGGGGCGAGGggacg---------CCCGACGGC----GTCCGAG
1020 1030 1040 1050 1060 1070 1080
1120 1130 1140 1150 1160 1170
seq1 GGGGcgacagaggcagcaGCAGCCCGAGGCCtgaggagaggagaccggcggcggcggca
|| ||| | || |||||||||||| || ||
seq2 GGCGcg------------GTGGCGCGAGGCCtgagggaggggacgcg------------
1090 1100 1110
CONREAL ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00253 | 50.00 | 27 | 34 | 32 | 39 | 1 | 5.016 | 4.131 | 0.90 | 0.86 | cap | C L | | M00101 | 51.35 | 28 | 34 | 33 | 39 | 2 | 3.667 | 5.021 | 0.84 | 0.90 | CdxA | C L | | M00498* | 50.00 | 28 | 35 | 33 | 40 | 1 | 6.258 | 5.394 | 0.90 | 0.87 | Pro only | C L | | M00496* | 50.00 | 28 | 35 | 33 | 40 | 1 | 6.723 | 4.244 | 0.92 | 0.81 | Pro only | C L | | M00101 | 51.35 | 29 | 35 | 34 | 40 | 1 | 3.820 | 4.579 | 0.85 | 0.88 | CdxA | C L | | M00100 | 51.35 | 29 | 35 | 34 | 40 | 1 | 4.191 | 6.269 | 0.81 | 0.92 | CdxA | C L | | M00101 | 51.35 | 30 | 36 | 35 | 41 | 1 | 5.861 | 3.676 | 0.93 | 0.84 | CdxA | C L | | M00101 | 54.05 | 42 | 48 | 113 | 119 | 1 | 3.591 | 2.982 | 0.84 | 0.82 | CdxA | C only | | M00734* | 51.28 | 42 | 50 | 113 | 121 | 1 | 6.386 | 6.020 | 0.83 | 0.82 | Pro only | C only | | MA0039 | 50.00 | 43 | 52 | 114 | 123 | 1 | 5.166 | 6.186 | 0.81 | 0.84 | Gklf | C only | | M00101 | 54.05 | 44 | 50 | 115 | 121 | 1 | 4.263 | 2.982 | 0.87 | 0.82 | CdxA | C only | | M00318* | 52.63 | 44 | 51 | 115 | 122 | 1 | 7.558 | 7.558 | 0.83 | 0.83 | Pro only | C only | | M00395 | 51.28 | 44 | 52 | 115 | 123 | 2 | 3.659 | 3.757 | 0.80 | 0.81 | HOXA3 | C only | | M00101 | 54.05 | 45 | 51 | 116 | 122 | 1 | 3.743 | 2.902 | 0.85 | 0.81 | CdxA | C only | | M00704* | 50.00 | 80 | 85 | 182 | 187 | 1 | 5.117 | 5.117 | 0.82 | 0.82 | Pro only | C only | | M00726* | 52.78 | 150 | 155 | 204 | 209 | 1 | 4.409 | 8.133 | 0.80 | 1.00 | Pro only | C only | | M00148 | 51.35 | 157 | 163 | 296 | 302 | 1 | 5.572 | 8.540 | 0.86 | 0.98 | SRY | C only | | M00975* | 51.28 | 157 | 165 | 296 | 304 | 2 | 7.390 | 5.269 | 0.90 | 0.82 | Pro only | C only | | M00712* | 52.63 | 158 | 165 | 297 | 304 | 1 | 7.066 | 6.002 | 0.86 | 0.82 | Pro only | C only | | M00003 | 52.50 | 158 | 167 | 297 | 306 | 1 | 7.183 | 8.751 | 0.81 | 0.86 | v-Myb | C only | | M00183 | 52.50 | 158 | 167 | 297 | 306 | 1 | 7.584 | 5.869 | 0.89 | 0.82 | c-Myb | C only | | M00253 | 55.26 | 159 | 166 | 298 | 305 | 2 | 4.744 | 6.247 | 0.89 | 0.95 | cap | C only | | MA0103 | 55.56 | 160 | 165 | 299 | 304 | 1 | 4.562 | 4.562 | 0.81 | 0.81 | deltaEF1 | C only | | MA0100 | 55.26 | 160 | 167 | 299 | 306 | 2 | 6.238 | 6.260 | 0.84 | 0.84 | c-MYB_1 | C only | | M00913* | 53.85 | 160 | 168 | 299 | 307 | 1 | 6.575 | 7.262 | 0.88 | 0.91 | Pro only | C only | | M00773* | 51.22 | 160 | 170 | 299 | 309 | 2 | 6.491 | 7.509 | 0.84 | 0.88 | Pro only | C only | | MA0081 | 54.05 | 368 | 374 | 310 | 316 | 1 | 9.933 | 8.051 | 0.98 | 0.92 | SPI-B | C only | | M00658* | 52.63 | 368 | 375 | 310 | 317 | 1 | 9.944 | 8.029 | 0.94 | 0.88 | Pro only | C only | | M00108 | 52.50 | 368 | 377 | 310 | 319 | 1 | 10.912 | 8.908 | 0.91 | 0.84 | NRF-2 | C only | | MA0062 | 52.50 | 368 | 377 | 310 | 319 | 1 | 10.912 | 8.908 | 0.91 | 0.84 | NRF-2 | C only | | MA0039 | 52.50 | 368 | 377 | 310 | 319 | 1 | 5.939 | 9.916 | 0.83 | 0.95 | Gklf | C only | | M00771* | 54.76 | 369 | 380 | 311 | 322 | 2 | 7.359 | 7.397 | 0.80 | 0.80 | Pro only | C only | | M00971* | 52.63 | 369 | 376 | 311 | 318 | 2 | 8.080 | 8.590 | 0.89 | 0.91 | Pro only | C only | | MA0080 | 52.78 | 370 | 375 | 312 | 317 | 1 | 8.711 | 8.184 | 1.00 | 0.98 | SPI-1 | C only | | MA0098 | 52.78 | 370 | 375 | 312 | 317 | 2 | 7.797 | 7.633 | 1.00 | 0.99 | c-ETS | C only | | M00500* | 52.63 | 370 | 377 | 312 | 319 | 2 | 5.902 | 8.565 | 0.87 | 0.97 | Pro only | C only | | M00498* | 52.63 | 370 | 377 | 312 | 319 | 2 | 4.668 | 5.533 | 0.85 | 0.88 | Pro only | C only | | M00499* | 52.63 | 370 | 377 | 312 | 319 | 2 | 5.727 | 6.150 | 0.89 | 0.91 | Pro only | C only | | M00496* | 52.63 | 370 | 377 | 312 | 319 | 2 | 5.438 | 4.798 | 0.86 | 0.84 | Pro only | C only | | M00497* | 52.63 | 370 | 377 | 312 | 319 | 2 | 6.977 | 6.196 | 0.95 | 0.91 | Pro only | C only | | M00655* | 51.35 | 370 | 376 | 312 | 318 | 2 | 6.124 | 6.628 | 0.84 | 0.86 | Pro only | C only | | MA0057 | 55.00 | 371 | 380 | 313 | 322 | 1 | 6.086 | 5.671 | 0.81 | 0.80 | MZF_5-13 | C only | | M00704* | 52.78 | 371 | 376 | 313 | 318 | 1 | 5.117 | 5.117 | 0.82 | 0.82 | Pro only | C only | | MA0057 | 55.00 | 372 | 381 | 314 | 323 | 1 | 5.809 | 7.104 | 0.80 | 0.85 | MZF_5-13 | C only | | M00986* | 61.11 | 374 | 379 | 316 | 321 | 1 | 5.768 | 6.177 | 0.90 | 0.92 | Pro only | C only | | MA0039 | 57.50 | 374 | 383 | 316 | 325 | 1 | 5.647 | 5.186 | 0.82 | 0.81 | Gklf | C only | | MA0098 | 55.56 | 382 | 387 | 324 | 329 | 2 | 5.977 | 3.658 | 0.92 | 0.81 | c-ETS | C only | | M00396 | 51.35 | 384 | 390 | 326 | 332 | 2 | 4.748 | 5.116 | 0.87 | 0.89 | En-1 | C only | | M00101 | 54.05 | 385 | 391 | 327 | 333 | 1 | 2.960 | 4.263 | 0.82 | 0.87 | CdxA | C only | | M00253 | 50.00 | 410 | 417 | 399 | 406 | 2 | 4.707 | 2.751 | 0.89 | 0.81 | cap | C only | | MA0109 | 52.63 | 433 | 440 | 449 | 456 | 2 | 5.458 | 4.984 | 0.84 | 0.82 | RUSH1-alfa | C L M | | M00183 | 55.00 | 434 | 443 | 450 | 459 | 1 | 6.097 | 7.393 | 0.83 | 0.89 | c-Myb | C L M | | M00253 | 57.89 | 435 | 442 | 451 | 458 | 2 | 4.778 | 5.231 | 0.89 | 0.91 | cap | C L M | | M00773* | 60.98 | 436 | 446 | 452 | 462 | 2 | 5.978 | 6.491 | 0.82 | 0.84 | Pro only | C L M | | M00913* | 58.97 | 436 | 444 | 452 | 460 | 1 | 6.278 | 6.925 | 0.87 | 0.90 | Pro only | C L M | | M00630* | 58.97 | 437 | 445 | 453 | 461 | 1 | 6.263 | 5.430 | 0.85 | 0.81 | Pro only | C L M | | M00253 | 60.53 | 440 | 447 | 456 | 463 | 2 | 2.912 | 4.165 | 0.81 | 0.87 | cap | C L M | | M00805* | 61.11 | 442 | 447 | 458 | 463 | 2 | 7.016 | 5.716 | 0.88 | 0.83 | Pro only | C L M | | M00253 | 68.42 | 450 | 457 | 466 | 473 | 2 | 5.576 | 5.599 | 0.93 | 0.93 | cap | C L M | | M00314* | 71.05 | 452 | 459 | 468 | 475 | 2 | 5.867 | 5.235 | 0.87 | 0.84 | Pro only | C L M | | M00313* | 71.05 | 452 | 459 | 468 | 475 | 2 | 5.673 | 5.523 | 0.87 | 0.86 | Pro only | C L M | | M00315* | 71.05 | 452 | 459 | 468 | 475 | 2 | 5.596 | 5.312 | 0.87 | 0.85 | Pro only | C L M | | M00418 | 65.85 | 452 | 462 | 468 | 478 | 2 | 7.107 | 7.535 | 0.80 | 0.82 | TGIF | C L M | | MA0103 | 61.11 | 458 | 463 | 474 | 479 | 2 | 6.503 | 5.313 | 0.89 | 0.84 | deltaEF1 | C L M | | M00912* | 66.67 | 459 | 470 | 475 | 486 | 2 | 7.437 | 8.807 | 0.85 | 0.89 | Pro only | C L M | | M00770* | 66.67 | 459 | 470 | 475 | 486 | 1 | 6.311 | 7.236 | 0.84 | 0.87 | Pro only | C L M | | M00109 | 63.64 | 459 | 472 | 475 | 488 | 1 | 8.580 | 9.782 | 0.87 | 0.90 | C/EBPbeta | C L M | | M00690* | 65.79 | 460 | 467 | 476 | 483 | 2 | 5.504 | 6.201 | 0.84 | 0.86 | Pro only | C L M | | M00962* | 64.10 | 463 | 471 | 479 | 487 | 1 | 5.464 | 6.392 | 0.83 | 0.87 | Pro only | C L M | | M00500* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.383 | 6.011 | 0.86 | 0.88 | Pro only | C L M | | M00498* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.233 | 6.097 | 0.87 | 0.90 | Pro only | C L M | | MA0109 | 65.79 | 464 | 471 | 480 | 487 | 2 | 7.153 | 8.196 | 0.91 | 0.96 | RUSH1-alfa | C L M | | M00499* | 65.79 | 464 | 471 | 480 | 487 | 2 | 6.782 | 7.681 | 0.93 | 0.97 | Pro only | C L M | | M00493* | 65.79 | 464 | 471 | 480 | 487 | 2 | 6.131 | 5.814 | 0.88 | 0.87 | Pro only | C L M | | M00496* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.225 | 6.731 | 0.86 | 0.92 | Pro only | C L M | | M00494* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.439 | 4.578 | 0.84 | 0.81 | Pro only | C L M | | M00497* | 65.79 | 464 | 471 | 480 | 487 | 2 | 4.146 | 4.451 | 0.82 | 0.83 | Pro only | C L M | | M00962* | 66.67 | 465 | 473 | 481 | 489 | 1 | 5.479 | 4.922 | 0.83 | 0.81 | Pro only | C L M | | M00921* | 65.79 | 465 | 472 | 481 | 488 | 2 | 5.738 | 6.200 | 0.83 | 0.85 | Pro only | C L M | | M00734* | 69.23 | 466 | 474 | 482 | 490 | 1 | 11.921 | 6.429 | 1.00 | 0.83 | Pro only | C L M | | M00445* | 65.12 | 466 | 478 | 482 | 494 | 2 | 7.391 | 6.501 | 0.83 | 0.80 | Pro only | C L M | | M00377 | 68.29 | 467 | 477 | 483 | 493 | 1 | 7.503 | 7.414 | 0.84 | 0.84 | Pax-4 | C L M | | M00770* | 50.00 | 495 | 506 | 586 | 597 | 1 | 5.300 | 9.539 | 0.80 | 0.95 | Pro only | C only | | M00975* | 51.28 | 500 | 508 | 591 | 599 | 2 | 7.117 | 9.148 | 0.89 | 0.96 | Pro only | C only | | MA0067 | 50.00 | 503 | 510 | 594 | 601 | 1 | 3.998 | 3.998 | 0.80 | 0.80 | Pax-2 | C only | | MA0095 | 50.00 | 506 | 511 | 597 | 602 | 2 | 6.225 | 6.225 | 0.89 | 0.89 | Yin-Yang | C only | | M00964* | 50.00 | 552 | 563 | 609 | 620 | 1 | 5.159 | 5.578 | 0.80 | 0.81 | Pro only | C L M | | M00921* | 52.63 | 553 | 560 | 610 | 617 | 2 | 5.595 | 5.595 | 0.83 | 0.83 | Pro only | C L M | | M00428* | 52.63 | 553 | 560 | 610 | 617 | 2 | 3.952 | 3.952 | 0.81 | 0.81 | Pro only | C L M | | M00962* | 51.28 | 553 | 561 | 610 | 618 | 1 | 5.773 | 5.280 | 0.85 | 0.83 | Pro only | C L M | | M00986* | 50.00 | 553 | 558 | 610 | 615 | 1 | 4.105 | 4.105 | 0.82 | 0.82 | Pro only | C L M | | M00646* | 52.63 | 554 | 561 | 611 | 618 | 1 | 7.680 | 8.150 | 0.89 | 0.91 | Pro only | C L M | | M00716* | 52.63 | 554 | 561 | 611 | 618 | 1 | 5.838 | 5.974 | 0.83 | 0.83 | Pro only | C L M | | M00921* | 55.26 | 555 | 562 | 612 | 619 | 2 | 6.399 | 6.861 | 0.86 | 0.88 | Pro only | C L M | | M00428* | 57.89 | 565 | 572 | 622 | 629 | 1 | 4.009 | 4.511 | 0.81 | 0.83 | Pro only | C L M | | M00986* | 52.78 | 570 | 575 | 627 | 632 | 1 | 4.223 | 5.706 | 0.83 | 0.90 | Pro only | C L M | | M00646* | 55.26 | 571 | 578 | 628 | 635 | 1 | 6.725 | 10.433 | 0.85 | 1.00 | Pro only | C L | | M00986* | 52.78 | 571 | 576 | 628 | 633 | 1 | 5.430 | 4.918 | 0.89 | 0.86 | Pro only | C L | | M00159 | 58.14 | 582 | 594 | 648 | 660 | 1 | 7.611 | 5.220 | 0.88 | 0.80 | C/EBP | C L M | | M00083 | 52.63 | 582 | 589 | 648 | 655 | 1 | 6.396 | 6.628 | 0.82 | 0.83 | MZF1 | C L M | | M00486* | 61.54 | 586 | 594 | 652 | 660 | 1 | 4.872 | 5.183 | 0.84 | 0.85 | Pro only | C L M | | MA0032 | 63.16 | 587 | 594 | 653 | 660 | 1 | 7.209 | 6.986 | 0.99 | 0.97 | FREAC-3 | C L M | | M00750* | 62.16 | 587 | 593 | 653 | 659 | 1 | 8.420 | 6.801 | 0.95 | 0.89 | Pro only | C L M | | M00148 | 64.86 | 589 | 595 | 655 | 661 | 1 | 6.998 | 6.998 | 0.92 | 0.92 | SRY | C L M | | MA0075 | 68.57 | 590 | 594 | 656 | 660 | 1 | 4.766 | 4.766 | 0.82 | 0.82 | S8 | C L M | | MA0080 | 66.67 | 591 | 596 | 657 | 662 | 1 | 4.391 | 4.391 | 0.82 | 0.82 | SPI-1 | C L M | | M00690* | 68.42 | 597 | 604 | 663 | 670 | 2 | 6.091 | 6.473 | 0.86 | 0.87 | Pro only | C L M | | M00974* | 63.41 | 599 | 609 | 665 | 675 | 1 | 7.650 | 7.242 | 0.84 | 0.83 | Pro only | C L M | | M00419 | 61.90 | 601 | 612 | 667 | 678 | 1 | 4.742 | 5.378 | 0.80 | 0.82 | MEIS1 | C L M | | MA0109 | 65.79 | 603 | 610 | 669 | 676 | 2 | 5.123 | 5.729 | 0.83 | 0.85 | RUSH1-alfa | C L M | | M00792* | 64.10 | 604 | 612 | 670 | 678 | 1 | 6.619 | 5.856 | 0.90 | 0.86 | Pro only | C L M | | MA0100 | 63.16 | 605 | 612 | 671 | 678 | 1 | 6.317 | 6.401 | 0.84 | 0.84 | c-MYB_1 | C L M | | M00500* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.976 | 4.838 | 0.88 | 0.84 | Pro only | C L M | | M00498* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.533 | 4.668 | 0.88 | 0.85 | Pro only | C L M | | M00499* | 57.89 | 759 | 766 | 762 | 769 | 1 | 7.211 | 6.788 | 0.95 | 0.93 | Pro only | C L M | | M00496* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.395 | 6.035 | 0.86 | 0.89 | Pro only | C L M | | M00497* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.899 | 6.836 | 0.90 | 0.94 | Pro only | C L M | | MA0080 | 55.56 | 761 | 766 | 764 | 769 | 2 | 6.483 | 8.711 | 0.91 | 1.00 | SPI-1 | C L M | | MA0098 | 55.56 | 761 | 766 | 764 | 769 | 1 | 6.300 | 7.797 | 0.93 | 1.00 | c-ETS | C L M | | M00486* | 58.97 | 771 | 779 | 774 | 782 | 1 | 5.834 | 6.213 | 0.88 | 0.90 | Pro only | C L M | | M00967* | 61.54 | 774 | 782 | 777 | 785 | 1 | 5.262 | 5.262 | 0.80 | 0.80 | Pro only | C L M | | M00809* | 58.14 | 774 | 786 | 777 | 789 | 2 | 7.916 | 9.284 | 0.80 | 0.83 | Pro only | C L M | | MA0041 | 57.14 | 775 | 786 | 778 | 789 | 2 | 7.971 | 7.289 | 0.83 | 0.81 | HFH-2 | C L M | | M00624* | 59.46 | 776 | 782 | 779 | 785 | 1 | 5.560 | 5.560 | 0.86 | 0.86 | Pro only | C L M | | M00395 | 56.41 | 788 | 796 | 795 | 803 | 2 | 4.235 | 3.816 | 0.83 | 0.81 | HOXA3 | C only | | M00486* | 56.41 | 789 | 797 | 796 | 804 | 1 | 5.069 | 5.272 | 0.85 | 0.86 | Pro only | C only | | M00971* | 55.26 | 793 | 800 | 800 | 807 | 2 | 5.207 | 6.952 | 0.80 | 0.86 | Pro only | C only | | MA0098 | 55.56 | 794 | 799 | 801 | 806 | 2 | 3.674 | 7.229 | 0.81 | 0.97 | c-ETS | C only | | M00498* | 60.53 | 806 | 813 | 814 | 821 | 2 | 5.094 | 7.226 | 0.86 | 0.94 | Pro only | C only | | M00496* | 60.53 | 806 | 813 | 814 | 821 | 2 | 4.963 | 6.105 | 0.84 | 0.89 | Pro only | C only | | M00100 | 59.46 | 806 | 812 | 814 | 820 | 2 | 5.476 | 5.215 | 0.88 | 0.87 | CdxA | C only | | M00486* | 58.97 | 806 | 814 | 814 | 822 | 1 | 4.977 | 5.178 | 0.84 | 0.85 | Pro only | C only | | M00925* | 61.54 | 807 | 815 | 815 | 823 | 2 | 7.916 | 6.056 | 0.88 | 0.82 | Pro only | C L M | | M00462* | 60.00 | 807 | 816 | 815 | 824 | 2 | 5.777 | 6.024 | 0.85 | 0.86 | Pro only | C L M | | MA0038 | 57.50 | 808 | 817 | 816 | 825 | 1 | 8.434 | 7.798 | 0.88 | 0.86 | Gfi | C L M | | M00396 | 59.46 | 809 | 815 | 817 | 823 | 2 | 3.803 | 3.803 | 0.81 | 0.81 | En-1 | C L M | | MA0037 | 58.33 | 809 | 814 | 817 | 822 | 2 | 5.162 | 5.162 | 0.86 | 0.86 | GATA-3 | C L M | | MA0035 | 58.33 | 809 | 814 | 817 | 822 | 2 | 4.265 | 4.265 | 0.85 | 0.85 | GATA-1 | C L M | | MA0036 | 60.00 | 810 | 814 | 818 | 822 | 2 | 4.000 | 4.000 | 0.85 | 0.85 | GATA-2 | C L M | | M00148 | 59.46 | 810 | 816 | 818 | 824 | 1 | 5.526 | 5.526 | 0.86 | 0.86 | SRY | C L M | | M00630* | 56.41 | 810 | 818 | 818 | 826 | 2 | 5.260 | 5.260 | 0.81 | 0.81 | Pro only | C L M | | M00462* | 55.00 | 810 | 819 | 818 | 827 | 2 | 4.645 | 4.645 | 0.81 | 0.81 | Pro only | C L M | | M00076 | 55.00 | 810 | 819 | 818 | 827 | 2 | 4.377 | 4.377 | 0.81 | 0.81 | GATA-2 | C L M | | MA0095 | 58.33 | 811 | 816 | 819 | 824 | 1 | 5.123 | 5.123 | 0.83 | 0.83 | Yin-Yang | C L M | | MA0089 | 58.33 | 811 | 816 | 819 | 824 | 2 | 6.811 | 6.811 | 0.91 | 0.91 | TCF11-MafG | C L M | | M00624* | 56.76 | 811 | 817 | 819 | 825 | 1 | 4.524 | 4.524 | 0.81 | 0.81 | Pro only | C L M | | M00285 | 51.16 | 811 | 823 | 819 | 831 | 1 | 7.570 | 6.869 | 0.88 | 0.85 | TCF11 | C L M | | MA0035 | 58.33 | 812 | 817 | 820 | 825 | 2 | 5.498 | 5.498 | 0.91 | 0.91 | GATA-1 | C L M | | M00655* | 56.76 | 812 | 818 | 820 | 826 | 1 | 7.813 | 7.813 | 0.91 | 0.91 | Pro only | C L M | | M00971* | 55.26 | 812 | 819 | 820 | 827 | 1 | 5.834 | 5.834 | 0.82 | 0.82 | Pro only | C L M | | M00253 | 55.26 | 812 | 819 | 820 | 827 | 1 | 6.400 | 6.400 | 0.96 | 0.96 | cap | C L M | | MA0036 | 60.00 | 813 | 817 | 821 | 825 | 2 | 5.978 | 5.978 | 0.96 | 0.96 | GATA-2 | C L M | | MA0098 | 58.33 | 813 | 818 | 821 | 826 | 1 | 7.229 | 7.229 | 0.97 | 0.97 | c-ETS | C L M | | M00743* | 60.53 | 821 | 828 | 829 | 836 | 1 | 6.503 | 7.812 | 0.82 | 0.87 | Pro only | C L M | | M00986* | 50.00 | 903 | 908 | 866 | 871 | 2 | 3.593 | 4.481 | 0.80 | 0.84 | Pro only | C L | | M00500* | 52.63 | 918 | 925 | 897 | 904 | 2 | 7.969 | 4.413 | 0.95 | 0.82 | Pro only | C M | | M00498* | 52.63 | 918 | 925 | 897 | 904 | 2 | 5.233 | 4.369 | 0.87 | 0.83 | Pro only | C M | | MA0080 | 50.00 | 918 | 923 | 897 | 902 | 1 | 8.579 | 4.897 | 0.99 | 0.84 | SPI-1 | C M | | M00491* | 60.47 | 922 | 934 | 901 | 913 | 2 | 8.041 | 11.563 | 0.80 | 0.88 | Pro only | C L M | | M00986* | 55.56 | 923 | 928 | 902 | 907 | 2 | 6.401 | 3.834 | 0.93 | 0.81 | Pro only | C L M | | MA0079 | 60.00 | 924 | 933 | 903 | 912 | 2 | 6.570 | 8.598 | 0.82 | 0.89 | SP1 | C L M | | M00720* | 61.54 | 925 | 933 | 904 | 912 | 2 | 7.530 | 8.270 | 0.84 | 0.87 | Pro only | C L M | | M00986* | 61.11 | 928 | 933 | 907 | 912 | 2 | 5.289 | 4.722 | 0.88 | 0.85 | Pro only | C L M | | M00431* | 63.16 | 933 | 940 | 912 | 919 | 1 | 6.288 | 4.994 | 0.85 | 0.81 | Pro only | C L M | | M00428* | 63.16 | 933 | 940 | 912 | 919 | 1 | 5.146 | 4.685 | 0.85 | 0.84 | Pro only | C L M | | M00695* | 56.76 | 937 | 943 | 916 | 922 | 2 | 7.007 | 5.823 | 0.89 | 0.84 | Pro only | C L M | | M00428* | 50.00 | 942 | 949 | 921 | 928 | 2 | 4.736 | 6.218 | 0.84 | 0.90 | Pro only | C L M | | M00803* | 50.00 | 945 | 950 | 924 | 929 | 2 | 4.808 | 4.808 | 0.81 | 0.81 | Pro only | C L M | | M00253 | 65.79 | 959 | 966 | 942 | 949 | 2 | 3.745 | 4.104 | 0.85 | 0.86 | cap | C L M | | M00468* | 64.86 | 959 | 965 | 942 | 948 | 2 | 7.266 | 6.440 | 0.94 | 0.89 | Pro only | C L M | | M00253 | 68.42 | 960 | 967 | 943 | 950 | 1 | 3.122 | 3.609 | 0.82 | 0.84 | cap | C L M | | M00470* | 69.23 | 961 | 969 | 944 | 952 | 2 | 6.454 | 6.454 | 0.85 | 0.85 | Pro only | C L M | | M00469* | 69.23 | 961 | 969 | 944 | 952 | 2 | 6.341 | 6.341 | 0.87 | 0.87 | Pro only | C L M | | MA0003 | 69.23 | 961 | 969 | 944 | 952 | 2 | 6.341 | 6.341 | 0.87 | 0.87 | AP2alpha | C L M | | M00314* | 68.42 | 961 | 968 | 944 | 951 | 2 | 5.867 | 5.867 | 0.87 | 0.87 | Pro only | C L M | | M00313* | 68.42 | 961 | 968 | 944 | 951 | 2 | 5.673 | 5.673 | 0.87 | 0.87 | Pro only | C L M | | M00315* | 68.42 | 961 | 968 | 944 | 951 | 2 | 5.596 | 5.596 | 0.87 | 0.87 | Pro only | C L M | | M00634* | 73.81 | 962 | 973 | 945 | 956 | 2 | 7.057 | 7.057 | 0.80 | 0.80 | Pro only | C L M | | MA0002 | 71.79 | 962 | 970 | 945 | 953 | 1 | 6.844 | 6.844 | 0.84 | 0.84 | AML-1 | C L M | | MA0048 | 73.81 | 963 | 974 | 946 | 957 | 2 | 8.917 | 8.917 | 0.82 | 0.82 | Hen-1 | C L M | | M00277 | 73.81 | 963 | 974 | 946 | 957 | 2 | 6.885 | 6.885 | 0.81 | 0.81 | Lmo2 complex | C L M | | M00271 | 75.00 | 964 | 969 | 947 | 952 | 1 | 5.416 | 5.416 | 0.83 | 0.83 | AML-1a | C L M | | M00253 | 76.32 | 965 | 972 | 948 | 955 | 2 | 5.465 | 5.465 | 0.92 | 0.92 | cap | C L M | | M00644* | 75.68 | 965 | 971 | 948 | 954 | 2 | 6.451 | 6.451 | 0.85 | 0.85 | Pro only | C L M | | M00695* | 75.68 | 965 | 971 | 948 | 954 | 1 | 6.047 | 6.047 | 0.85 | 0.85 | Pro only | C L M | | M00644* | 75.68 | 966 | 972 | 949 | 955 | 1 | 8.574 | 8.574 | 0.94 | 0.94 | Pro only | C L M | | M00803* | 75.00 | 970 | 975 | 953 | 958 | 2 | 4.932 | 6.310 | 0.82 | 0.88 | Pro only | C L M | | M00915* | 69.77 | 976 | 988 | 959 | 971 | 2 | 9.784 | 9.050 | 0.88 | 0.86 | Pro only | C L M | | M00470* | 74.36 | 977 | 985 | 960 | 968 | 1 | 8.151 | 7.624 | 0.90 | 0.89 | Pro only | C L M | | M00469* | 74.36 | 977 | 985 | 960 | 968 | 1 | 8.284 | 8.284 | 0.93 | 0.93 | Pro only | C L M | | MA0003 | 74.36 | 977 | 985 | 960 | 968 | 1 | 8.284 | 8.284 | 0.93 | 0.93 | AP2alpha | C L M | | M00322* | 72.50 | 977 | 986 | 960 | 969 | 1 | 6.101 | 7.399 | 0.81 | 0.85 | Pro only | C L M | | M00428* | 68.42 | 980 | 987 | 963 | 970 | 1 | 4.727 | 4.130 | 0.84 | 0.81 | Pro only | C L M | | M00803* | 69.44 | 981 | 986 | 964 | 969 | 1 | 8.895 | 8.895 | 1.00 | 1.00 | Pro only | C L M | | M00803* | 69.44 | 981 | 986 | 964 | 969 | 2 | 5.517 | 5.517 | 0.85 | 0.85 | Pro only | C L M | | M00976* | 64.10 | 981 | 989 | 964 | 972 | 1 | 5.614 | 5.961 | 0.80 | 0.82 | Pro only | C L M | | M00322* | 62.50 | 981 | 990 | 964 | 973 | 1 | 9.545 | 7.606 | 0.91 | 0.85 | Pro only | C only | | M00322* | 62.50 | 981 | 990 | 964 | 973 | 2 | 7.753 | 8.141 | 0.86 | 0.87 | Pro only | C only | | M00716* | 63.16 | 982 | 989 | 965 | 972 | 1 | 6.700 | 10.047 | 0.86 | 1.00 | Pro only | C L M | | M00803* | 63.89 | 983 | 988 | 966 | 971 | 1 | 7.517 | 7.517 | 0.94 | 0.94 | Pro only | C L M | | M00803* | 63.89 | 983 | 988 | 966 | 971 | 2 | 7.517 | 7.517 | 0.94 | 0.94 | Pro only | C L M | | M00716* | 57.89 | 991 | 998 | 992 | 999 | 1 | 6.966 | 7.579 | 0.87 | 0.90 | Pro only | C only | | M00803* | 58.33 | 992 | 997 | 993 | 998 | 1 | 8.243 | 8.895 | 0.97 | 1.00 | Pro only | C only | | M00428* | 57.89 | 993 | 1000 | 994 | 1001 | 2 | 7.680 | 4.459 | 0.95 | 0.83 | Pro only | C only | | M00720* | 66.67 | 1007 | 1015 | 1008 | 1016 | 1 | 11.368 | 8.182 | 0.98 | 0.87 | Pro only | C only | | M00253 | 65.79 | 1007 | 1014 | 1008 | 1015 | 2 | 5.186 | 5.897 | 0.91 | 0.94 | cap | C only | | M00695* | 64.86 | 1007 | 1013 | 1008 | 1014 | 1 | 5.779 | 5.779 | 0.84 | 0.84 | Pro only | C only | | M00271 | 63.89 | 1012 | 1017 | 1013 | 1018 | 1 | 5.832 | 5.416 | 0.84 | 0.83 | AML-1a | C only | | M00469* | 61.54 | 1015 | 1023 | 1016 | 1024 | 2 | 5.823 | 6.265 | 0.85 | 0.87 | Pro only | C only | | MA0003 | 61.54 | 1015 | 1023 | 1016 | 1024 | 2 | 5.823 | 6.265 | 0.85 | 0.87 | AP2alpha | C only | | M00695* | 59.46 | 1019 | 1025 | 1020 | 1026 | 1 | 5.823 | 9.770 | 0.84 | 1.00 | Pro only | C only | | M00803* | 58.33 | 1021 | 1026 | 1022 | 1027 | 1 | 8.895 | 6.243 | 1.00 | 0.88 | Pro only | C only | | M00803* | 58.33 | 1021 | 1026 | 1022 | 1027 | 2 | 5.517 | 4.573 | 0.85 | 0.80 | Pro only | C only | | M00695* | 62.16 | 1024 | 1030 | 1025 | 1031 | 1 | 9.770 | 5.663 | 1.00 | 0.84 | Pro only | C only | | M00803* | 58.33 | 1026 | 1031 | 1027 | 1032 | 1 | 6.243 | 4.864 | 0.88 | 0.82 | Pro only | C only | | M00976* | 51.28 | 1063 | 1071 | 1045 | 1053 | 1 | 6.654 | 5.558 | 0.84 | 0.80 | Pro only | C only | | M00196 | 51.16 | 1063 | 1075 | 1045 | 1057 | 1 | 9.459 | 7.921 | 0.84 | 0.80 | Sp1 | C only | | M00716* | 50.00 | 1064 | 1071 | 1046 | 1053 | 1 | 5.638 | 7.579 | 0.82 | 0.90 | Pro only | C only | | M00931* | 50.00 | 1065 | 1074 | 1047 | 1056 | 1 | 8.365 | 7.623 | 0.83 | 0.81 | Pro only | C only | | M00470* | 51.28 | 1066 | 1074 | 1048 | 1056 | 2 | 5.322 | 8.308 | 0.81 | 0.91 | Pro only | C only | | M00469* | 51.28 | 1066 | 1074 | 1048 | 1056 | 2 | 6.135 | 8.783 | 0.86 | 0.95 | Pro only | C only | | MA0003 | 51.28 | 1066 | 1074 | 1048 | 1056 | 2 | 6.135 | 8.783 | 0.86 | 0.95 | AP2alpha | C only | | M00803* | 50.00 | 1067 | 1072 | 1049 | 1054 | 1 | 4.543 | 6.864 | 0.80 | 0.91 | Pro only | C only | | M00428* | 52.63 | 1068 | 1075 | 1050 | 1057 | 2 | 3.866 | 3.866 | 0.80 | 0.80 | Pro only | C only | | M00986* | 52.78 | 1069 | 1074 | 1051 | 1056 | 1 | 7.013 | 7.013 | 0.96 | 0.96 | Pro only | C only | | M00695* | 56.76 | 1070 | 1076 | 1052 | 1058 | 1 | 5.555 | 5.555 | 0.83 | 0.83 | Pro only | C only | | M00803* | 55.56 | 1070 | 1075 | 1052 | 1057 | 1 | 4.808 | 4.808 | 0.81 | 0.81 | Pro only | C only | | M00986* | 55.56 | 1070 | 1075 | 1052 | 1057 | 1 | 4.722 | 4.722 | 0.85 | 0.85 | Pro only | C only | | M00649* | 55.26 | 1070 | 1077 | 1052 | 1059 | 1 | 8.155 | 8.774 | 0.85 | 0.88 | Pro only | C only | | M00716* | 55.26 | 1071 | 1078 | 1053 | 1060 | 1 | 6.700 | 7.102 | 0.86 | 0.88 | Pro only | C only | | M00803* | 52.78 | 1072 | 1077 | 1054 | 1059 | 1 | 8.895 | 5.436 | 1.00 | 0.84 | Pro only | C only | | M00469* | 51.28 | 1093 | 1101 | 1066 | 1074 | 2 | 5.729 | 8.905 | 0.85 | 0.95 | Pro only | C only | | MA0003 | 51.28 | 1093 | 1101 | 1066 | 1074 | 2 | 5.729 | 8.905 | 0.85 | 0.95 | AP2alpha | C only | | M00470* | 53.85 | 1106 | 1114 | 1075 | 1083 | 1 | 5.137 | 5.452 | 0.81 | 0.82 | Pro only | C only | | M00986* | 50.00 | 1111 | 1116 | 1080 | 1085 | 1 | 6.177 | 4.481 | 0.92 | 0.84 | Pro only | C only | | M00915* | 65.12 | 1131 | 1143 | 1088 | 1100 | 1 | 9.475 | 6.894 | 0.87 | 0.80 | Pro only | C L M | | M00428* | 63.16 | 1134 | 1141 | 1091 | 1098 | 2 | 5.065 | 6.548 | 0.85 | 0.91 | Pro only | C L M | | M00470* | 61.54 | 1134 | 1142 | 1091 | 1099 | 1 | 10.915 | 5.291 | 0.99 | 0.81 | Pro only | C L M | | M00470* | 61.54 | 1134 | 1142 | 1091 | 1099 | 2 | 10.419 | 7.835 | 0.97 | 0.89 | Pro only | C L M | | M00469* | 61.54 | 1134 | 1142 | 1091 | 1099 | 1 | 10.105 | 4.313 | 0.99 | 0.80 | Pro only | C L M | | M00469* | 61.54 | 1134 | 1142 | 1091 | 1099 | 2 | 9.691 | 8.192 | 0.98 | 0.93 | Pro only | C L M | | MA0003 | 61.54 | 1134 | 1142 | 1091 | 1099 | 1 | 10.105 | 4.313 | 0.99 | 0.80 | AP2alpha | C L M | | MA0003 | 61.54 | 1134 | 1142 | 1091 | 1099 | 2 | 9.691 | 8.192 | 0.98 | 0.93 | AP2alpha | C L M |
LAGAN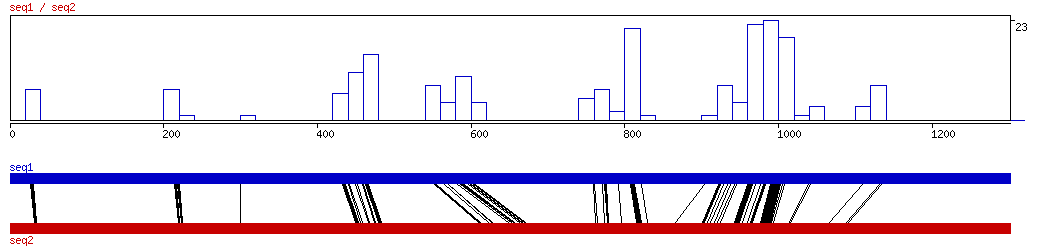
LAGAN ALIGNMENT 10 20 30 40 50 60 70 80 90
seq1 c-ttgagcaaattgg----tggatggtggtgTCATTTACTGaaacggaaaatacaggatgagcagattaaaggtgaacatgcaatgaatgaaattttcaa
||| | || | || | ||||||||| || || || ||| | | || ||||||| |||
seq2 tactgaacctcttataaccttgaccatcttgTCATTTAAAAaagagggtgtcaccctggtagccagtc-----ttaaagcaaaatgaatcctcattttgg
10 20 30 40 50 60 70 80 90
100 110 120 130 140 150 160 170 180 190
seq1 ttttgtgtatgttggtctgagatgccttttggatacatgttgtatacatgcttacatgagtaagcaactggtatgaattgacggagagggaggaaggaca
| || | | | | || || || | | | | || | |||| | || ||| | || || | |
seq2 aagtctgcactcaaggaaaaaaaaaagcatgactaaatatccaac---ttcctagctcagtagtcgtctttcctgagt------------agaaaaggct
100 110 120 130 140 150 160 170 180
200 210 220 230 240 250 260 270
seq1 ggg--------------------cagaagatagagctgTGGGAGTCATCGGTTCCTtgacagaatttacagcgtggtaatggatagactctatgctatcc
|| || | || |||| || |||| | | | || | | | || || | | || | ||
seq2 cggagtgttttaatccccttttccacgtggaagggctgGGGATGTCACCCACTGCCagagtgcttgtctagtatgccacaaaggatgcttgagatgtccc
190 200 210 220 230 240 250 260 270 280
280 290 300 310 320 330 340 350 360
seq1 atctatggaacaacacatctggagAAGAGAGaagagctggggaggctggactgtagccttgggagtggctattaaaataggaga---------------a
|| ||||| | | || |||| || | | || | | | | | | || ||||| || |
seq2 g-----gggacaacgcggtgaaacAACTGAGaggaaaggaagggggcaagcaaaaac---agaggagtaccctacaatagatgacggctagactagccca
290 300 310 320 330 340 350 360 370
370 380 390 400 410 420 430 440 450
seq1 ggggtggagcggaagggagagaggaactatgga----gtgcttagcaagagaagaaagtgttttaaggatggggatGATAAACTGCATTGAaaGCCACTG
||| | || | | ||| | | | || | || | | ||||| | ||| | | ||| |||||| ||| ||||
seq2 gggaaagtgcatagagaggaggaggagtctgtgcacgtagttttgggacagaagggatgaaaggaagaaaaaaggtGAGCAACTGCTCTGAggATAACTG
380 390 400 410 420 430 440 450 460 470
460 470 480 490 500 510 520 530 540
seq1 AGAGGTGGAGAAAAAATCAGGGcagagaagtaataaaaggcatcagcaacatggaagtcactggtgactcgcgaaaggcgatcac---------------
| |||| ||||||| || ||| |||| |||| | | || ||| || || | | | |||| | |
seq2 AAAGGTTGAGAAAAGATTGACGcacagaa----taaagagggttggctcagtgggagagccttggtatgcaagaaaccgagttccacccccacaaggggg
480 490 500 510 520 530 540 550 560 5
550 560 570 580 590 600
seq1 -------------------ctttggtg-----------tgTAGGGCACAAAAgCCTGGGGGGGTAAAg---------agAGAGTGGAAAGTAAGAATGTC
|||||| || |||||||| || || ||||| | |||| | | |||||||| | |
seq2 cagaggaggggaaacatggctttggcaacatggagagatgAAGGGCACAGAAaTTTGACGGGGTCAGtcaaagatctagAGTGGGCAAAGTAAGGGTTTG
70 580 590 600 610 620 630 640 650 660 6
610 620 630 640 650 660 670 680 690 700
seq1 GAGACAGTCGactattattgccattttatgaatgaagaaaccgaggctcaatatttgactgtctcattcaaggtgaaagccagtggggtcagatcctgga
||||||| || || | | || | || |
seq2 GAGACAGGTGatcatcaccgaca-------------------------------------------------------------------aaccccaata
70 680 690 700
710 720 730 740 750 760 770 780 790 8
seq1 cctagaatccatgcaatgattt----gtgcacatcgcctattgaagagagagatccaattCTCTTCCAaaacGTAAAAGCCAACACTTccctcAAAGGAG
| || | | | ||| || | | | | | | | | |||| || ||||||| | ||||||||| || |||| |
seq2 cggagtaccaaagcagtgttgttatagggaggcgcagccacccaggagattcccgcagcgCTCTTCCGacctTCGAAAGCCAACCAACccttcAACTAAA
710 720 730 740 750 760 770 780 790 800
00 810 820 830 840 850 860 870 880 890
seq1 Gcttcgag-----TTAAATCATCCTTTCACGATGCcgcttccattttaaagaaaaaaaaaaaaaaacgagaaggcctggtgcgggataggaatgagccga
| | || | ||||||||||| || || || | | || |||| |
seq2 GgatgcaggggccTGAAATCATCCTTCCAAGAAGCagtcataaccttggagaa---------------------------------------------gg
810 820 830 840 850
900 910 920 930 940 950 960 970
seq1 ggagctactGGCCCTctcggtcggggg----------------aAGACCCGCCCCTTTTCCGCAGCCCGAGCC----cggcgcagTCCACTGCGGCTGCG
||| | ||||| || | | | | ||| ||| ||| | ||| || |||||| | | || |||||||||||||
seq2 ccggcttccCGCCCTgtctgcagagcgggcgagcgcgctcgcgaAGGCCCCGCCCCTCGGCGCCGCGCGAGCCtcccctgagcgcCGCACTGCGGCTGCG
860 870 880 890 900 910 920 930 940 950
980 990 1000 1010 1020 1030 1040 1050 1060 1070
seq1 CGAGCCTGGCGCGCGC---GGGGCGGGAACGCGGAAGGGGCTGgggttgcggcgcggcggcgaggaccAGACCGGGGGCGGGgccggtagtgggagtgcg
| ||| |||||||| |||||||| |||||||||| || | |||| | | | | ||| ||||| || | | | ||
seq2 CCGGCCGGGCGCGCGGaggGGGGCGGGGACGCGGAAGGCGCGG-----------cggccggggaggctGCGGCGGCGGCGGCgcggtgaagcgagagccg
960 970 980 990 1000 1010 1020 1030 1040
1080 1090 1100 1110 1120 1130 1140 1150 1160 1170
seq1 gggcgcgcggtgacagcgcggggttggcggcgtgggacCCAGGGGGCgacagaggcagcaGCAGCCCGAGGCCtgaggagaggagaccggcggcggcggc
||||| || || | || || | |||| | |||| || | || |||||||||||| || | ||
seq2 acgcgcggggcgaggggacg------------------CCCGACGGCgtccgagggcgcgGTGGCGCGAGGCCtgagggaggg------------gacgc
1050 1060 1070 1080 1090 1100 1110
seq1 a
seq2 g
LAGAN ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00253 | 50.00 | 27 | 34 | 32 | 39 | 1 | 5.016 | 4.131 | 0.90 | 0.86 | cap | L C | | M00101 | 51.35 | 28 | 34 | 33 | 39 | 2 | 3.667 | 5.021 | 0.84 | 0.90 | CdxA | L C | | M00496* | 50.00 | 28 | 35 | 33 | 40 | 1 | 6.723 | 4.244 | 0.92 | 0.81 | Pro only | L C | | M00498* | 50.00 | 28 | 35 | 33 | 40 | 1 | 6.258 | 5.394 | 0.90 | 0.87 | Pro only | L C | | M00100 | 51.35 | 29 | 35 | 34 | 40 | 1 | 4.191 | 6.269 | 0.81 | 0.92 | CdxA | L C | | M00101 | 51.35 | 29 | 35 | 34 | 40 | 1 | 3.820 | 4.579 | 0.85 | 0.88 | CdxA | L C | | M00101 | 51.35 | 30 | 36 | 35 | 41 | 1 | 5.861 | 3.676 | 0.93 | 0.84 | CdxA | L C | | M00172 | 51.22 | 214 | 224 | 219 | 229 | 2 | 9.355 | 7.878 | 0.90 | 0.85 | AP-1 | L M | | M00113 | 50.00 | 215 | 226 | 220 | 231 | 2 | 8.574 | 9.671 | 0.85 | 0.88 | CREB | L M | | M00926* | 50.00 | 216 | 223 | 221 | 228 | 1 | 9.460 | 5.599 | 0.93 | 0.80 | Pro only | L M | | M00801* | 52.78 | 218 | 223 | 223 | 228 | 1 | 5.678 | 6.558 | 0.86 | 0.89 | Pro only | L M | | MA0067 | 52.63 | 218 | 225 | 223 | 230 | 1 | 4.623 | 5.060 | 0.83 | 0.84 | Pax-2 | L M | | M00285 | 53.49 | 219 | 231 | 224 | 236 | 1 | 7.355 | 6.447 | 0.87 | 0.84 | TCF11 | L M | | MA0089 | 52.78 | 219 | 224 | 224 | 229 | 2 | 8.692 | 5.919 | 0.99 | 0.87 | TCF11-MafG | L M | | M00253 | 52.63 | 220 | 227 | 225 | 232 | 1 | 4.688 | 5.267 | 0.89 | 0.91 | cap | L M | | M00747* | 51.35 | 300 | 306 | 300 | 306 | 2 | 5.658 | 6.075 | 0.81 | 0.82 | Pro only | L B | | MA0109 | 52.63 | 433 | 440 | 449 | 456 | 2 | 5.458 | 4.984 | 0.84 | 0.82 | RUSH1-alfa | L C M | | M00183 | 55.00 | 434 | 443 | 450 | 459 | 1 | 6.097 | 7.393 | 0.83 | 0.89 | c-Myb | L C M | | M00253 | 57.89 | 435 | 442 | 451 | 458 | 2 | 4.778 | 5.231 | 0.89 | 0.91 | cap | L C M | | M00773* | 60.98 | 436 | 446 | 452 | 462 | 2 | 5.978 | 6.491 | 0.82 | 0.84 | Pro only | L C M | | M00913* | 58.97 | 436 | 444 | 452 | 460 | 1 | 6.278 | 6.925 | 0.87 | 0.90 | Pro only | L C M | | M00630* | 58.97 | 437 | 445 | 453 | 461 | 1 | 6.263 | 5.430 | 0.85 | 0.81 | Pro only | L C M | | M00253 | 60.53 | 440 | 447 | 456 | 463 | 2 | 2.912 | 4.165 | 0.81 | 0.87 | cap | L C M | | M00805* | 61.11 | 442 | 447 | 458 | 463 | 2 | 7.016 | 5.716 | 0.88 | 0.83 | Pro only | L C M | | M00253 | 68.42 | 450 | 457 | 466 | 473 | 2 | 5.576 | 5.599 | 0.93 | 0.93 | cap | L C M | | M00313* | 71.05 | 452 | 459 | 468 | 475 | 2 | 5.673 | 5.523 | 0.87 | 0.86 | Pro only | L C M | | M00314* | 71.05 | 452 | 459 | 468 | 475 | 2 | 5.867 | 5.235 | 0.87 | 0.84 | Pro only | L C M | | M00315* | 71.05 | 452 | 459 | 468 | 475 | 2 | 5.596 | 5.312 | 0.87 | 0.85 | Pro only | L C M | | M00418 | 65.85 | 452 | 462 | 468 | 478 | 2 | 7.107 | 7.535 | 0.80 | 0.82 | TGIF | L C M | | MA0103 | 61.11 | 458 | 463 | 474 | 479 | 2 | 6.503 | 5.313 | 0.89 | 0.84 | deltaEF1 | L C M | | M00109 | 63.64 | 459 | 472 | 475 | 488 | 1 | 8.580 | 9.782 | 0.87 | 0.90 | C/EBPbeta | L C M | | M00770* | 66.67 | 459 | 470 | 475 | 486 | 1 | 6.311 | 7.236 | 0.84 | 0.87 | Pro only | L C M | | M00912* | 66.67 | 459 | 470 | 475 | 486 | 2 | 7.437 | 8.807 | 0.85 | 0.89 | Pro only | L C M | | M00690* | 65.79 | 460 | 467 | 476 | 483 | 2 | 5.504 | 6.201 | 0.84 | 0.86 | Pro only | L C M | | M00962* | 64.10 | 463 | 471 | 479 | 487 | 1 | 5.464 | 6.392 | 0.83 | 0.87 | Pro only | L C M | | M00493* | 65.79 | 464 | 471 | 480 | 487 | 2 | 6.131 | 5.814 | 0.88 | 0.87 | Pro only | L C M | | M00494* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.439 | 4.578 | 0.84 | 0.81 | Pro only | L C M | | M00496* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.225 | 6.731 | 0.86 | 0.92 | Pro only | L C M | | M00497* | 65.79 | 464 | 471 | 480 | 487 | 2 | 4.146 | 4.451 | 0.82 | 0.83 | Pro only | L C M | | M00498* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.233 | 6.097 | 0.87 | 0.90 | Pro only | L C M | | M00499* | 65.79 | 464 | 471 | 480 | 487 | 2 | 6.782 | 7.681 | 0.93 | 0.97 | Pro only | L C M | | M00500* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.383 | 6.011 | 0.86 | 0.88 | Pro only | L C M | | MA0109 | 65.79 | 464 | 471 | 480 | 487 | 2 | 7.153 | 8.196 | 0.91 | 0.96 | RUSH1-alfa | L C M | | M00921* | 65.79 | 465 | 472 | 481 | 488 | 2 | 5.738 | 6.200 | 0.83 | 0.85 | Pro only | L C M | | M00962* | 66.67 | 465 | 473 | 481 | 489 | 1 | 5.479 | 4.922 | 0.83 | 0.81 | Pro only | L C M | | M00445* | 65.12 | 466 | 478 | 482 | 494 | 2 | 7.391 | 6.501 | 0.83 | 0.80 | Pro only | L C M | | M00734* | 69.23 | 466 | 474 | 482 | 490 | 1 | 11.921 | 6.429 | 1.00 | 0.83 | Pro only | L C M | | M00377 | 68.29 | 467 | 477 | 483 | 493 | 1 | 7.503 | 7.414 | 0.84 | 0.84 | Pax-4 | L C M | | M00964* | 50.00 | 552 | 563 | 609 | 620 | 1 | 5.159 | 5.578 | 0.80 | 0.81 | Pro only | L C M | | M00428* | 52.63 | 553 | 560 | 610 | 617 | 2 | 3.952 | 3.952 | 0.81 | 0.81 | Pro only | L C M | | M00921* | 52.63 | 553 | 560 | 610 | 617 | 2 | 5.595 | 5.595 | 0.83 | 0.83 | Pro only | L C M | | M00962* | 51.28 | 553 | 561 | 610 | 618 | 1 | 5.773 | 5.280 | 0.85 | 0.83 | Pro only | L C M | | M00986* | 50.00 | 553 | 558 | 610 | 615 | 1 | 4.105 | 4.105 | 0.82 | 0.82 | Pro only | L C M | | M00646* | 52.63 | 554 | 561 | 611 | 618 | 1 | 7.680 | 8.150 | 0.89 | 0.91 | Pro only | L C M | | M00716* | 52.63 | 554 | 561 | 611 | 618 | 1 | 5.838 | 5.974 | 0.83 | 0.83 | Pro only | L C M | | M00921* | 55.26 | 555 | 562 | 612 | 619 | 2 | 6.399 | 6.861 | 0.86 | 0.88 | Pro only | L C M | | M00428* | 57.89 | 565 | 572 | 622 | 629 | 1 | 4.009 | 4.511 | 0.81 | 0.83 | Pro only | L C M | | M00986* | 52.78 | 570 | 575 | 627 | 632 | 1 | 4.223 | 5.706 | 0.83 | 0.90 | Pro only | L C M | | M00646* | 55.26 | 571 | 578 | 628 | 635 | 1 | 6.725 | 10.433 | 0.85 | 1.00 | Pro only | L C | | M00986* | 52.78 | 571 | 576 | 628 | 633 | 1 | 5.430 | 4.918 | 0.89 | 0.86 | Pro only | L C | | M00083 | 52.63 | 582 | 589 | 648 | 655 | 1 | 6.396 | 6.628 | 0.82 | 0.83 | MZF1 | L C M | | M00159 | 58.14 | 582 | 594 | 648 | 660 | 1 | 7.611 | 5.220 | 0.88 | 0.80 | C/EBP | L C M | | M00486* | 61.54 | 586 | 594 | 652 | 660 | 1 | 4.872 | 5.183 | 0.84 | 0.85 | Pro only | L C M | | M00750* | 62.16 | 587 | 593 | 653 | 659 | 1 | 8.420 | 6.801 | 0.95 | 0.89 | Pro only | L C M | | MA0032 | 63.16 | 587 | 594 | 653 | 660 | 1 | 7.209 | 6.986 | 0.99 | 0.97 | FREAC-3 | L C M | | M00148 | 64.86 | 589 | 595 | 655 | 661 | 1 | 6.998 | 6.998 | 0.92 | 0.92 | SRY | L C M | | MA0075 | 68.57 | 590 | 594 | 656 | 660 | 1 | 4.766 | 4.766 | 0.82 | 0.82 | S8 | L C M | | MA0080 | 66.67 | 591 | 596 | 657 | 662 | 1 | 4.391 | 4.391 | 0.82 | 0.82 | SPI-1 | L C M | | M00690* | 68.42 | 597 | 604 | 663 | 670 | 2 | 6.091 | 6.473 | 0.86 | 0.87 | Pro only | L C M | | M00974* | 63.41 | 599 | 609 | 665 | 675 | 1 | 7.650 | 7.242 | 0.84 | 0.83 | Pro only | L C M | | M00419 | 61.90 | 601 | 612 | 667 | 678 | 1 | 4.742 | 5.378 | 0.80 | 0.82 | MEIS1 | L C M | | MA0109 | 65.79 | 603 | 610 | 669 | 676 | 2 | 5.123 | 5.729 | 0.83 | 0.85 | RUSH1-alfa | L C M | | M00792* | 64.10 | 604 | 612 | 670 | 678 | 1 | 6.619 | 5.856 | 0.90 | 0.86 | Pro only | L C M | | MA0100 | 63.16 | 605 | 612 | 671 | 678 | 1 | 6.317 | 6.401 | 0.84 | 0.84 | c-MYB_1 | L C M | | M00496* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.395 | 6.035 | 0.86 | 0.89 | Pro only | L C M | | M00497* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.899 | 6.836 | 0.90 | 0.94 | Pro only | L C M | | M00498* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.533 | 4.668 | 0.88 | 0.85 | Pro only | L C M | | M00499* | 57.89 | 759 | 766 | 762 | 769 | 1 | 7.211 | 6.788 | 0.95 | 0.93 | Pro only | L C M | | M00500* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.976 | 4.838 | 0.88 | 0.84 | Pro only | L C M | | MA0080 | 55.56 | 761 | 766 | 764 | 769 | 2 | 6.483 | 8.711 | 0.91 | 1.00 | SPI-1 | L C M | | MA0098 | 55.56 | 761 | 766 | 764 | 769 | 1 | 6.300 | 7.797 | 0.93 | 1.00 | c-ETS | L C M | | M00486* | 58.97 | 771 | 779 | 774 | 782 | 1 | 5.834 | 6.213 | 0.88 | 0.90 | Pro only | L C M | | M00809* | 58.14 | 774 | 786 | 777 | 789 | 2 | 7.916 | 9.284 | 0.80 | 0.83 | Pro only | L C M | | M00967* | 61.54 | 774 | 782 | 777 | 785 | 1 | 5.262 | 5.262 | 0.80 | 0.80 | Pro only | L C M | | MA0041 | 57.14 | 775 | 786 | 778 | 789 | 2 | 7.971 | 7.289 | 0.83 | 0.81 | HFH-2 | L C M | | M00624* | 59.46 | 776 | 782 | 779 | 785 | 1 | 5.560 | 5.560 | 0.86 | 0.86 | Pro only | L C M | | M00313* | 50.00 | 792 | 799 | 795 | 802 | 2 | 5.108 | 4.395 | 0.84 | 0.81 | Pro only | L M | | M00315* | 50.00 | 792 | 799 | 795 | 802 | 2 | 4.900 | 4.234 | 0.83 | 0.80 | Pro only | L M | | M00462* | 60.00 | 807 | 816 | 815 | 824 | 2 | 5.777 | 6.024 | 0.85 | 0.86 | Pro only | L C M | | M00925* | 61.54 | 807 | 815 | 815 | 823 | 2 | 7.916 | 6.056 | 0.88 | 0.82 | Pro only | L C M | | MA0038 | 57.50 | 808 | 817 | 816 | 825 | 1 | 8.434 | 7.798 | 0.88 | 0.86 | Gfi | L C M | | M00396 | 59.46 | 809 | 815 | 817 | 823 | 2 | 3.803 | 3.803 | 0.81 | 0.81 | En-1 | L C M | | MA0035 | 58.33 | 809 | 814 | 817 | 822 | 2 | 4.265 | 4.265 | 0.85 | 0.85 | GATA-1 | L C M | | MA0037 | 58.33 | 809 | 814 | 817 | 822 | 2 | 5.162 | 5.162 | 0.86 | 0.86 | GATA-3 | L C M | | M00076 | 55.00 | 810 | 819 | 818 | 827 | 2 | 4.377 | 4.377 | 0.81 | 0.81 | GATA-2 | L C M | | M00148 | 59.46 | 810 | 816 | 818 | 824 | 1 | 5.526 | 5.526 | 0.86 | 0.86 | SRY | L C M | | M00462* | 55.00 | 810 | 819 | 818 | 827 | 2 | 4.645 | 4.645 | 0.81 | 0.81 | Pro only | L C M | | M00630* | 56.41 | 810 | 818 | 818 | 826 | 2 | 5.260 | 5.260 | 0.81 | 0.81 | Pro only | L C M | | MA0036 | 60.00 | 810 | 814 | 818 | 822 | 2 | 4.000 | 4.000 | 0.85 | 0.85 | GATA-2 | L C M | | M00285 | 51.16 | 811 | 823 | 819 | 831 | 1 | 7.570 | 6.869 | 0.88 | 0.85 | TCF11 | L C M | | M00624* | 56.76 | 811 | 817 | 819 | 825 | 1 | 4.524 | 4.524 | 0.81 | 0.81 | Pro only | L C M | | MA0089 | 58.33 | 811 | 816 | 819 | 824 | 2 | 6.811 | 6.811 | 0.91 | 0.91 | TCF11-MafG | L C M | | MA0095 | 58.33 | 811 | 816 | 819 | 824 | 1 | 5.123 | 5.123 | 0.83 | 0.83 | Yin-Yang | L C M | | M00253 | 55.26 | 812 | 819 | 820 | 827 | 1 | 6.400 | 6.400 | 0.96 | 0.96 | cap | L C M | | M00655* | 56.76 | 812 | 818 | 820 | 826 | 1 | 7.813 | 7.813 | 0.91 | 0.91 | Pro only | L C M | | M00971* | 55.26 | 812 | 819 | 820 | 827 | 1 | 5.834 | 5.834 | 0.82 | 0.82 | Pro only | L C M | | MA0035 | 58.33 | 812 | 817 | 820 | 825 | 2 | 5.498 | 5.498 | 0.91 | 0.91 | GATA-1 | L C M | | MA0036 | 60.00 | 813 | 817 | 821 | 825 | 2 | 5.978 | 5.978 | 0.96 | 0.96 | GATA-2 | L C M | | MA0098 | 58.33 | 813 | 818 | 821 | 826 | 1 | 7.229 | 7.229 | 0.97 | 0.97 | c-ETS | L C M | | M00743* | 60.53 | 821 | 828 | 829 | 836 | 1 | 6.503 | 7.812 | 0.82 | 0.87 | Pro only | L C M | | M00986* | 50.00 | 903 | 908 | 866 | 871 | 2 | 3.593 | 4.481 | 0.80 | 0.84 | Pro only | L C | | M00491* | 60.47 | 922 | 934 | 901 | 913 | 2 | 8.041 | 11.563 | 0.80 | 0.88 | Pro only | L C M | | M00986* | 55.56 | 923 | 928 | 902 | 907 | 2 | 6.401 | 3.834 | 0.93 | 0.81 | Pro only | L C M | | MA0079 | 60.00 | 924 | 933 | 903 | 912 | 2 | 6.570 | 8.598 | 0.82 | 0.89 | SP1 | L C M | | M00720* | 61.54 | 925 | 933 | 904 | 912 | 2 | 7.530 | 8.270 | 0.84 | 0.87 | Pro only | L C M | | M00986* | 61.11 | 928 | 933 | 907 | 912 | 2 | 5.289 | 4.722 | 0.88 | 0.85 | Pro only | L C M | | M00428* | 63.16 | 933 | 940 | 912 | 919 | 1 | 5.146 | 4.685 | 0.85 | 0.84 | Pro only | L C M | | M00431* | 63.16 | 933 | 940 | 912 | 919 | 1 | 6.288 | 4.994 | 0.85 | 0.81 | Pro only | L C M | | M00695* | 56.76 | 937 | 943 | 916 | 922 | 2 | 7.007 | 5.823 | 0.89 | 0.84 | Pro only | L C M | | M00428* | 50.00 | 942 | 949 | 921 | 928 | 2 | 4.736 | 6.218 | 0.84 | 0.90 | Pro only | L C M | | M00803* | 50.00 | 945 | 950 | 924 | 929 | 2 | 4.808 | 4.808 | 0.81 | 0.81 | Pro only | L C M | | M00253 | 65.79 | 959 | 966 | 942 | 949 | 2 | 3.745 | 4.104 | 0.85 | 0.86 | cap | L C M | | M00468* | 64.86 | 959 | 965 | 942 | 948 | 2 | 7.266 | 6.440 | 0.94 | 0.89 | Pro only | L C M | | M00253 | 68.42 | 960 | 967 | 943 | 950 | 1 | 3.122 | 3.609 | 0.82 | 0.84 | cap | L C M | | M00313* | 68.42 | 961 | 968 | 944 | 951 | 2 | 5.673 | 5.673 | 0.87 | 0.87 | Pro only | L C M | | M00314* | 68.42 | 961 | 968 | 944 | 951 | 2 | 5.867 | 5.867 | 0.87 | 0.87 | Pro only | L C M | | M00315* | 68.42 | 961 | 968 | 944 | 951 | 2 | 5.596 | 5.596 | 0.87 | 0.87 | Pro only | L C M | | M00469* | 69.23 | 961 | 969 | 944 | 952 | 2 | 6.341 | 6.341 | 0.87 | 0.87 | Pro only | L C M | | M00470* | 69.23 | 961 | 969 | 944 | 952 | 2 | 6.454 | 6.454 | 0.85 | 0.85 | Pro only | L C M | | MA0003 | 69.23 | 961 | 969 | 944 | 952 | 2 | 6.341 | 6.341 | 0.87 | 0.87 | AP2alpha | L C M | | M00634* | 73.81 | 962 | 973 | 945 | 956 | 2 | 7.057 | 7.057 | 0.80 | 0.80 | Pro only | L C M | | MA0002 | 71.79 | 962 | 970 | 945 | 953 | 1 | 6.844 | 6.844 | 0.84 | 0.84 | AML-1 | L C M | | M00277 | 73.81 | 963 | 974 | 946 | 957 | 2 | 6.885 | 6.885 | 0.81 | 0.81 | Lmo2 complex | L C M | | MA0048 | 73.81 | 963 | 974 | 946 | 957 | 2 | 8.917 | 8.917 | 0.82 | 0.82 | Hen-1 | L C M | | M00271 | 75.00 | 964 | 969 | 947 | 952 | 1 | 5.416 | 5.416 | 0.83 | 0.83 | AML-1a | L C M | | M00253 | 76.32 | 965 | 972 | 948 | 955 | 2 | 5.465 | 5.465 | 0.92 | 0.92 | cap | L C M | | M00644* | 75.68 | 965 | 971 | 948 | 954 | 2 | 6.451 | 6.451 | 0.85 | 0.85 | Pro only | L C M | | M00695* | 75.68 | 965 | 971 | 948 | 954 | 1 | 6.047 | 6.047 | 0.85 | 0.85 | Pro only | L C M | | M00644* | 75.68 | 966 | 972 | 949 | 955 | 1 | 8.574 | 8.574 | 0.94 | 0.94 | Pro only | L C M | | M00803* | 75.00 | 970 | 975 | 953 | 958 | 2 | 4.932 | 6.310 | 0.82 | 0.88 | Pro only | L C M | | M00915* | 72.09 | 976 | 988 | 959 | 971 | 2 | 9.784 | 9.050 | 0.88 | 0.86 | Pro only | L C M | | M00322* | 75.00 | 977 | 986 | 960 | 969 | 1 | 6.101 | 7.399 | 0.81 | 0.85 | Pro only | L C M | | M00469* | 74.36 | 977 | 985 | 960 | 968 | 1 | 8.284 | 8.284 | 0.93 | 0.93 | Pro only | L C M | | M00470* | 74.36 | 977 | 985 | 960 | 968 | 1 | 8.151 | 7.624 | 0.90 | 0.89 | Pro only | L C M | | MA0003 | 74.36 | 977 | 985 | 960 | 968 | 1 | 8.284 | 8.284 | 0.93 | 0.93 | AP2alpha | L C M | | M00428* | 71.05 | 980 | 987 | 963 | 970 | 1 | 4.727 | 4.130 | 0.84 | 0.81 | Pro only | L C M | | M00803* | 72.22 | 981 | 986 | 964 | 969 | 1 | 8.895 | 8.895 | 1.00 | 1.00 | Pro only | L C M | | M00803* | 72.22 | 981 | 986 | 964 | 969 | 2 | 5.517 | 5.517 | 0.85 | 0.85 | Pro only | L C M | | M00976* | 69.23 | 981 | 989 | 964 | 972 | 1 | 5.614 | 5.961 | 0.80 | 0.82 | Pro only | L C M | | M00716* | 68.42 | 982 | 989 | 965 | 972 | 1 | 6.700 | 10.047 | 0.86 | 1.00 | Pro only | L C M | | M00803* | 66.67 | 983 | 988 | 966 | 971 | 1 | 7.517 | 7.517 | 0.94 | 0.94 | Pro only | L C M | | M00803* | 66.67 | 983 | 988 | 966 | 971 | 2 | 7.517 | 7.517 | 0.94 | 0.94 | Pro only | L C M | | M00008 | 62.50 | 990 | 999 | 976 | 985 | 1 | 9.109 | 11.207 | 0.90 | 0.97 | Sp1 | L M | | M00649* | 63.16 | 990 | 997 | 976 | 983 | 1 | 10.557 | 10.557 | 0.94 | 0.94 | Pro only | L M | | M00695* | 64.86 | 990 | 996 | 976 | 982 | 1 | 9.502 | 9.502 | 0.99 | 0.99 | Pro only | L M | | M00803* | 63.89 | 990 | 995 | 976 | 981 | 1 | 4.808 | 4.808 | 0.81 | 0.81 | Pro only | L M | | M00931* | 62.50 | 990 | 999 | 976 | 985 | 1 | 11.469 | 13.658 | 0.91 | 0.96 | Pro only | L M | | M00938* | 60.87 | 990 | 1005 | 976 | 991 | 1 | 9.502 | 9.002 | 0.83 | 0.82 | Pro only | L M | | M00986* | 63.89 | 990 | 995 | 976 | 981 | 1 | 4.722 | 4.722 | 0.85 | 0.85 | Pro only | L M | | MA0079 | 62.50 | 990 | 999 | 976 | 985 | 1 | 8.561 | 10.535 | 0.89 | 0.95 | SP1 | L M | | M00716* | 65.79 | 991 | 998 | 977 | 984 | 1 | 6.966 | 7.102 | 0.87 | 0.88 | Pro only | L M | | M00803* | 66.67 | 992 | 997 | 978 | 983 | 1 | 8.243 | 8.243 | 0.97 | 0.97 | Pro only | L M | | M00428* | 65.79 | 993 | 1000 | 979 | 986 | 2 | 7.680 | 3.958 | 0.95 | 0.81 | Pro only | L M | | M00333* | 65.12 | 994 | 1006 | 980 | 992 | 1 | 7.391 | 8.555 | 0.82 | 0.85 | Pro only | L M | | M00986* | 66.67 | 994 | 999 | 980 | 985 | 1 | 7.115 | 7.524 | 0.96 | 0.98 | Pro only | L M | | M00938* | 65.22 | 996 | 1011 | 982 | 997 | 1 | 9.979 | 8.685 | 0.84 | 0.81 | Pro only | L M | | M00716* | 68.42 | 997 | 1004 | 983 | 990 | 1 | 6.514 | 8.071 | 0.85 | 0.92 | Pro only | L M | | MA0028 | 70.00 | 998 | 1007 | 984 | 993 | 1 | 7.006 | 7.450 | 0.85 | 0.86 | Elk-1 | L M | | M00108 | 72.50 | 1000 | 1009 | 986 | 995 | 1 | 9.143 | 8.334 | 0.85 | 0.82 | NRF-2 | L M | | M00428* | 73.68 | 1000 | 1007 | 986 | 993 | 2 | 4.184 | 4.184 | 0.82 | 0.82 | Pro only | L M | | M00658* | 73.68 | 1000 | 1007 | 986 | 993 | 1 | 6.688 | 6.688 | 0.83 | 0.83 | Pro only | L M | | MA0062 | 72.50 | 1000 | 1009 | 986 | 995 | 1 | 9.143 | 8.334 | 0.85 | 0.82 | NRF-2 | L M | | MA0081 | 72.97 | 1000 | 1006 | 986 | 992 | 1 | 5.707 | 5.707 | 0.84 | 0.84 | SPI-B | L M | | M00695* | 75.68 | 1001 | 1007 | 987 | 993 | 1 | 5.454 | 5.454 | 0.83 | 0.83 | Pro only | L M | | M00971* | 73.68 | 1001 | 1008 | 987 | 994 | 2 | 8.080 | 8.080 | 0.89 | 0.89 | Pro only | L M | | M00492* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 8.448 | 7.226 | 0.91 | 0.87 | Pro only | L M | | M00496* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 5.438 | 5.517 | 0.86 | 0.87 | Pro only | L M | | M00497* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 6.977 | 7.065 | 0.95 | 0.95 | Pro only | L M | | M00498* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 4.668 | 5.233 | 0.85 | 0.87 | Pro only | L M | | M00499* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 5.727 | 5.685 | 0.89 | 0.89 | Pro only | L M | | M00500* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 5.902 | 6.673 | 0.87 | 0.90 | Pro only | L M | | M00655* | 72.97 | 1002 | 1008 | 988 | 994 | 2 | 6.124 | 6.124 | 0.84 | 0.84 | Pro only | L M | | MA0080 | 75.00 | 1002 | 1007 | 988 | 993 | 1 | 8.711 | 8.711 | 1.00 | 1.00 | SPI-1 | L M | | MA0098 | 75.00 | 1002 | 1007 | 988 | 993 | 2 | 7.797 | 7.797 | 1.00 | 1.00 | c-ETS | L M | | M00704* | 75.00 | 1003 | 1008 | 989 | 994 | 1 | 5.117 | 5.117 | 0.82 | 0.82 | Pro only | L M | | MA0103 | 75.00 | 1005 | 1010 | 991 | 996 | 2 | 4.459 | 5.629 | 0.81 | 0.86 | deltaEF1 | L M | | M00695* | 67.57 | 1007 | 1013 | 993 | 999 | 1 | 5.779 | 5.663 | 0.84 | 0.84 | Pro only | L M | | M00982* | 50.00 | 1039 | 1052 | 1014 | 1027 | 2 | 11.718 | 12.247 | 0.84 | 0.85 | Pro only | L only | | M00469* | 51.28 | 1041 | 1049 | 1016 | 1024 | 2 | 7.850 | 6.265 | 0.92 | 0.87 | Pro only | L only | | M00470* | 51.28 | 1041 | 1049 | 1016 | 1024 | 2 | 8.599 | 5.926 | 0.92 | 0.83 | Pro only | L only | | MA0003 | 51.28 | 1041 | 1049 | 1016 | 1024 | 2 | 7.850 | 6.265 | 0.92 | 0.87 | AP2alpha | L only | | M00469* | 53.85 | 1109 | 1117 | 1066 | 1074 | 2 | 7.016 | 8.905 | 0.89 | 0.95 | Pro only | L M | | M00470* | 53.85 | 1109 | 1117 | 1066 | 1074 | 2 | 7.718 | 8.735 | 0.89 | 0.92 | Pro only | L M | | MA0003 | 53.85 | 1109 | 1117 | 1066 | 1074 | 2 | 7.016 | 8.905 | 0.89 | 0.95 | AP2alpha | L M | | M00915* | 65.12 | 1131 | 1143 | 1088 | 1100 | 1 | 9.475 | 6.894 | 0.87 | 0.80 | Pro only | L C M | | M00428* | 63.16 | 1134 | 1141 | 1091 | 1098 | 2 | 5.065 | 6.548 | 0.85 | 0.91 | Pro only | L C M | | M00469* | 61.54 | 1134 | 1142 | 1091 | 1099 | 1 | 10.105 | 4.313 | 0.99 | 0.80 | Pro only | L C M | | M00469* | 61.54 | 1134 | 1142 | 1091 | 1099 | 2 | 9.691 | 8.192 | 0.98 | 0.93 | Pro only | L C M | | M00470* | 61.54 | 1134 | 1142 | 1091 | 1099 | 1 | 10.915 | 5.291 | 0.99 | 0.81 | Pro only | L C M | | M00470* | 61.54 | 1134 | 1142 | 1091 | 1099 | 2 | 10.419 | 7.835 | 0.97 | 0.89 | Pro only | L C M | | MA0003 | 61.54 | 1134 | 1142 | 1091 | 1099 | 1 | 10.105 | 4.313 | 0.99 | 0.80 | AP2alpha | L C M | | MA0003 | 61.54 | 1134 | 1142 | 1091 | 1099 | 2 | 9.691 | 8.192 | 0.98 | 0.93 | AP2alpha | L C M |
MAVID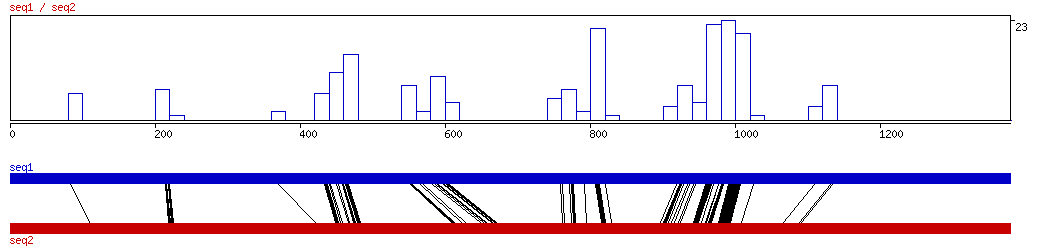
MAVID ALIGNMENT 10 20 30 40 50 60
seq1 ------------------cttgagcaaattggtggatggtggtgtcatttactgaaacggaaaatacaggatg----------------------agcag
||||| || | |||||||| |||| | ||| || ||||
seq2 tactgaacctcttataaccttgacca-------------tcttgtcattt----------aaaaaagagggtgtcaccctggtagccagtcttaaagcaa
10 20 30 40 50 60 70
70 80 90 100 110 120 130 140 150
seq1 attaaag---------gtgaacatgcaatgaATGAAATTttcaattttgtgtatgttggtctgagatgccttttggatacatgttgtatacatgcttaca
| | || | | |||| | || |||| || || || | || || || | ||
seq2 aatgaatcctcattttggaagtctgcactcaAGGAAAAAaaaaagcatgactaaat--------------------atccaactt---------cctagc
80 90 100 110 120 130 140 1
160 170 180 190 200 210 220
seq1 tgagtaagcaactggtatgaattga-------cggagagg------------------gaggaaggacagggcagaagatagagctgTGGGAGTCATCGG
| |||| | || ||| | || ||||| | | |||||| |||| || |||| |
seq2 tcagtagtcgtctttcctgagtagaaaaggctcggagtgttttaatccccttttccacgtggaagg-----------------gctgGGGATGTCACCCA
50 160 170 180 190 200 210 220 230
230 240 250 260 270 280 290 300
seq1 TTCCTtgacagaatttacagcgtggta--atggatagactctatgctatccatctatggaacaac--------acatctggagaagagag----------
| | || | | | || || | | |||| ||| || || ||||| ||| || |||| || ||
seq2 CTGCCagagtgcttgtctagtatgccacaaaggatgcttgagatg-------tcccggggacaacgcggtgaaacaact-gagaggaaaggaagggggca
240 250 260 270 280 290 300 310 320
310 320 330 340 350 360 3
seq1 ---aagagctggggag-------------------gctggactgtagccttgggagtggctattaaaataggagaaggggt----------------gga
|| | | | |||| ||| ||| ||||| |||| | || | | ||||| ||| || ||
seq2 agcaaaaacagaggagtaccctacaatagatgacggctagac--tagcccagggaaagtgcat--agagaggaggaggagtctgtgcacgtagttttggg
330 340 350 360 370 380 390 400 410 42
70 380 390 400 410 420 430 440 450 460
seq1 gCGGAAGgga-gagaggaactatggagtgcttagcaagagaagaaagtgttttaaggatggggatGATAAACTGCATTGAaaGCCACTGAGAGGTGGAGA
| ||||||| || |||| |||| ||| | ||| |||||| ||| ||||| |||| ||||
seq2 aCAGAAGggatgaaagga--------------------agaaaaaa----------------ggtGAGCAACTGCTCTGAggATAACTGAAAGGTTGAGA
0 430 440 450 460 470 480
470 480 490 500 510 520 530
seq1 AAAAATCAGGGcagagaagtaataaaaggcatcagcaacatggaagtcactggtgactcgcgaaa--------------------------------ggc
||| || ||| || |||||| | | || ||| || || | | | |||| ||
seq2 AAAGATTGACGcacag----aataaagagggttggctcagtgggagagccttggtatgcaagaaaccgagttccacccccacaagggggcagaggagggg
490 500 510 520 530 540 550 560 570 58
540 550 560 570 580 590 600 610
seq1 gatcac--ctttggtg-----------tgTAGGGCACAAAAgCCTGGGGGGGT-----aaaga----gAGAGTGGAAAGTAAGAATGTCGAGACAGTCGa
| || |||||| || |||||||| || || ||||| ||||| ||| | | |||||||| | | ||||||| ||
seq2 aaacatggctttggcaacatggagagatgAAGGGCACAGAAaTTTGACGGGGTcagtcaaagatctagAGTGGGCAAAGTAAGGGTTTGGAGACAGGTGa
0 590 600 610 620 630 640 650 660 670 68
620 630 640 650 660 670 680 690 700 710
seq1 ctattattgccattttatgaatgaagaaaccgaggctcaatatttgactgtctcattcaaggtgaaagccagtggggtcagatcctggacctagaatcca
|| | | | ||||| || || || | | |
seq2 tcatcaccg---------------acaaacc----------------------------------------------------ccaatacggagtaccaa
0 690 700 710
720 730 740 750 760 770 780 790 800
seq1 tgcaatgattt----gtgcacatcgcctattgaagagagagatccaattCTCTTCCAaaacGTAAAAGCCAACACTTccctcAAAGGAGGcttcgag---
||| || | | | | | | | | |||| || ||||||| | ||||||||| || |||| | | | ||
seq2 agcagtgttgttatagggaggcgcagccacccaggagattcccgcagcgCTCTTCCGacctTCGAAAGCCAACCAACccttcAACTAAAGgatgcagggg
720 730 740 750 760 770 780 790 800 810
810 820 830 840 850 860 870 880 890 900
seq1 --TTAAATCATCCTTTCACGATGCcgcttccattttaaagaaaaaaaaaaaaaaacgagaaggcctggtgcgggataggaatg---agccgaggagctac
| ||||||||||| || || || | | || ||||||||| | | | | | || ||| | ||||
seq2 ccTGAAATCATCCTTCCAAGAAGC-------------------agtcataaccttggagaaggccggcttcccgccctgtctgcagagcgggcgagc---
820 830 840 850 860 870 880 890
910 920 930 940 950 960 970 980 990
seq1 tggccctctcggtcggGGGAAGACCCGCCCCTTTTCCGCAGCCCGAGCC----cggcgcagTCCACTGCGGCTGCGCGAGCCTGGCGCGCGC---GGGGC
|| ||| | |||| ||| ||| | ||| || |||||| | | || |||||||||||||| ||| |||||||| |||||
seq2 --gcgctc--------GCGAAGGCCCCGCCCCTCGGCGCCGCGCGAGCCtcccctgagcgcCGCACTGCGGCTGCGCCGGCCGGGCGCGCGGaggGGGGC
900 910 920 930 940 950 960 970 980
1000 1010 1020 1030 1040 1050 1060 1070 1080 1090
seq1 GGGAACGCGGAAGGGGCTGGggttgcggcGCGGCGGcgaggaccagaccgggggcggggccggtagtgggagtgcggggcgcgcggtgacagcgcggggt
||| |||||||||| || | || ||| | ||| ||| ||| ||||| | |||| | ||| || | ||||||| | ||
seq2 GGGGACGCGGAAGGCGCGGCgg--ccgggGAGGCTGcg---------gcggcggcggcg-cggtgaagcgagagc-cgacgcgcggggcgag--------
990 1000 1010 1020 1030 1040 1050
1100 1110 1120 1130 1140 1150 1160 1170
seq1 tggcggcgtgggac-CCAGGGGGCgacagaggcagcaGCAGCCCGAGGCCtgaggagaggagaccggcggcggcggca
||||| || | |||| | |||| || | || |||||||||||| |||| ||| ||
seq2 ---------gggacgCCCGACGGCgtccgagggcgcgGTGGCGCGAGGCCtgagg-gaggggac-----------gcg
1060 1070 1080 1090 1100 1110
MAVID ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00493* | 50.00 | 83 | 90 | 109 | 116 | 2 | 5.295 | 7.366 | 0.84 | 0.94 | Pro only | M only | | M00494* | 50.00 | 83 | 90 | 109 | 116 | 2 | 6.333 | 7.613 | 0.88 | 0.94 | Pro only | M only | | M00496* | 50.00 | 83 | 90 | 109 | 116 | 2 | 4.909 | 5.901 | 0.84 | 0.88 | Pro only | M only | | M00498* | 50.00 | 83 | 90 | 109 | 116 | 2 | 3.699 | 6.097 | 0.81 | 0.90 | Pro only | M only | | M00499* | 50.00 | 83 | 90 | 109 | 116 | 2 | 4.068 | 6.563 | 0.82 | 0.93 | Pro only | M only | | M00500* | 50.00 | 83 | 90 | 109 | 116 | 2 | 6.699 | 6.746 | 0.90 | 0.90 | Pro only | M only | | M00172 | 51.22 | 214 | 224 | 219 | 229 | 2 | 9.355 | 7.878 | 0.90 | 0.85 | AP-1 | M L | | M00113 | 50.00 | 215 | 226 | 220 | 231 | 2 | 8.574 | 9.671 | 0.85 | 0.88 | CREB | M L | | M00926* | 50.00 | 216 | 223 | 221 | 228 | 1 | 9.460 | 5.599 | 0.93 | 0.80 | Pro only | M L | | M00801* | 52.78 | 218 | 223 | 223 | 228 | 1 | 5.678 | 6.558 | 0.86 | 0.89 | Pro only | M L | | MA0067 | 52.63 | 218 | 225 | 223 | 230 | 1 | 4.623 | 5.060 | 0.83 | 0.84 | Pax-2 | M L | | M00285 | 53.49 | 219 | 231 | 224 | 236 | 1 | 7.355 | 6.447 | 0.87 | 0.84 | TCF11 | M L | | MA0089 | 52.78 | 219 | 224 | 224 | 229 | 2 | 8.692 | 5.919 | 0.99 | 0.87 | TCF11-MafG | M L | | M00253 | 52.63 | 220 | 227 | 225 | 232 | 1 | 4.688 | 5.267 | 0.89 | 0.91 | cap | M L | | MA0080 | 50.00 | 370 | 375 | 421 | 426 | 1 | 8.711 | 5.628 | 1.00 | 0.87 | SPI-1 | M only | | MA0098 | 50.00 | 370 | 375 | 421 | 426 | 2 | 7.797 | 3.822 | 1.00 | 0.82 | c-ETS | M only | | MA0109 | 52.63 | 433 | 440 | 449 | 456 | 2 | 5.458 | 4.984 | 0.84 | 0.82 | RUSH1-alfa | M C L | | M00183 | 55.00 | 434 | 443 | 450 | 459 | 1 | 6.097 | 7.393 | 0.83 | 0.89 | c-Myb | M C L | | M00253 | 57.89 | 435 | 442 | 451 | 458 | 2 | 4.778 | 5.231 | 0.89 | 0.91 | cap | M C L | | M00773* | 60.98 | 436 | 446 | 452 | 462 | 2 | 5.978 | 6.491 | 0.82 | 0.84 | Pro only | M C L | | M00913* | 58.97 | 436 | 444 | 452 | 460 | 1 | 6.278 | 6.925 | 0.87 | 0.90 | Pro only | M C L | | M00630* | 58.97 | 437 | 445 | 453 | 461 | 1 | 6.263 | 5.430 | 0.85 | 0.81 | Pro only | M C L | | M00253 | 60.53 | 440 | 447 | 456 | 463 | 2 | 2.912 | 4.165 | 0.81 | 0.87 | cap | M C L | | M00805* | 61.11 | 442 | 447 | 458 | 463 | 2 | 7.016 | 5.716 | 0.88 | 0.83 | Pro only | M C L | | M00253 | 68.42 | 450 | 457 | 466 | 473 | 2 | 5.576 | 5.599 | 0.93 | 0.93 | cap | M C L | | M00313* | 71.05 | 452 | 459 | 468 | 475 | 2 | 5.673 | 5.523 | 0.87 | 0.86 | Pro only | M C L | | M00314* | 71.05 | 452 | 459 | 468 | 475 | 2 | 5.867 | 5.235 | 0.87 | 0.84 | Pro only | M C L | | M00315* | 71.05 | 452 | 459 | 468 | 475 | 2 | 5.596 | 5.312 | 0.87 | 0.85 | Pro only | M C L | | M00418 | 65.85 | 452 | 462 | 468 | 478 | 2 | 7.107 | 7.535 | 0.80 | 0.82 | TGIF | M C L | | MA0103 | 61.11 | 458 | 463 | 474 | 479 | 2 | 6.503 | 5.313 | 0.89 | 0.84 | deltaEF1 | M C L | | M00109 | 63.64 | 459 | 472 | 475 | 488 | 1 | 8.580 | 9.782 | 0.87 | 0.90 | C/EBPbeta | M C L | | M00770* | 66.67 | 459 | 470 | 475 | 486 | 1 | 6.311 | 7.236 | 0.84 | 0.87 | Pro only | M C L | | M00912* | 66.67 | 459 | 470 | 475 | 486 | 2 | 7.437 | 8.807 | 0.85 | 0.89 | Pro only | M C L | | M00690* | 65.79 | 460 | 467 | 476 | 483 | 2 | 5.504 | 6.201 | 0.84 | 0.86 | Pro only | M C L | | M00962* | 64.10 | 463 | 471 | 479 | 487 | 1 | 5.464 | 6.392 | 0.83 | 0.87 | Pro only | M C L | | M00493* | 65.79 | 464 | 471 | 480 | 487 | 2 | 6.131 | 5.814 | 0.88 | 0.87 | Pro only | M C L | | M00494* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.439 | 4.578 | 0.84 | 0.81 | Pro only | M C L | | M00496* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.225 | 6.731 | 0.86 | 0.92 | Pro only | M C L | | M00497* | 65.79 | 464 | 471 | 480 | 487 | 2 | 4.146 | 4.451 | 0.82 | 0.83 | Pro only | M C L | | M00498* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.233 | 6.097 | 0.87 | 0.90 | Pro only | M C L | | M00499* | 65.79 | 464 | 471 | 480 | 487 | 2 | 6.782 | 7.681 | 0.93 | 0.97 | Pro only | M C L | | M00500* | 65.79 | 464 | 471 | 480 | 487 | 2 | 5.383 | 6.011 | 0.86 | 0.88 | Pro only | M C L | | MA0109 | 65.79 | 464 | 471 | 480 | 487 | 2 | 7.153 | 8.196 | 0.91 | 0.96 | RUSH1-alfa | M C L | | M00921* | 65.79 | 465 | 472 | 481 | 488 | 2 | 5.738 | 6.200 | 0.83 | 0.85 | Pro only | M C L | | M00962* | 66.67 | 465 | 473 | 481 | 489 | 1 | 5.479 | 4.922 | 0.83 | 0.81 | Pro only | M C L | | M00445* | 65.12 | 466 | 478 | 482 | 494 | 2 | 7.391 | 6.501 | 0.83 | 0.80 | Pro only | M C L | | M00734* | 69.23 | 466 | 474 | 482 | 490 | 1 | 11.921 | 6.429 | 1.00 | 0.83 | Pro only | M C L | | M00377 | 68.29 | 467 | 477 | 483 | 493 | 1 | 7.503 | 7.414 | 0.84 | 0.84 | Pax-4 | M C L | | M00964* | 50.00 | 552 | 563 | 609 | 620 | 1 | 5.159 | 5.578 | 0.80 | 0.81 | Pro only | M C L | | M00428* | 52.63 | 553 | 560 | 610 | 617 | 2 | 3.952 | 3.952 | 0.81 | 0.81 | Pro only | M C L | | M00921* | 52.63 | 553 | 560 | 610 | 617 | 2 | 5.595 | 5.595 | 0.83 | 0.83 | Pro only | M C L | | M00962* | 51.28 | 553 | 561 | 610 | 618 | 1 | 5.773 | 5.280 | 0.85 | 0.83 | Pro only | M C L | | M00986* | 50.00 | 553 | 558 | 610 | 615 | 1 | 4.105 | 4.105 | 0.82 | 0.82 | Pro only | M C L | | M00646* | 52.63 | 554 | 561 | 611 | 618 | 1 | 7.680 | 8.150 | 0.89 | 0.91 | Pro only | M C L | | M00716* | 52.63 | 554 | 561 | 611 | 618 | 1 | 5.838 | 5.974 | 0.83 | 0.83 | Pro only | M C L | | M00921* | 55.26 | 555 | 562 | 612 | 619 | 2 | 6.399 | 6.861 | 0.86 | 0.88 | Pro only | M C L | | M00428* | 57.89 | 565 | 572 | 622 | 629 | 1 | 4.009 | 4.511 | 0.81 | 0.83 | Pro only | M C L | | M00986* | 52.78 | 570 | 575 | 627 | 632 | 1 | 4.223 | 5.706 | 0.83 | 0.90 | Pro only | M C L | | M00083 | 52.63 | 582 | 589 | 648 | 655 | 1 | 6.396 | 6.628 | 0.82 | 0.83 | MZF1 | M C L | | M00159 | 58.14 | 582 | 594 | 648 | 660 | 1 | 7.611 | 5.220 | 0.88 | 0.80 | C/EBP | M C L | | M00486* | 61.54 | 586 | 594 | 652 | 660 | 1 | 4.872 | 5.183 | 0.84 | 0.85 | Pro only | M C L | | M00750* | 62.16 | 587 | 593 | 653 | 659 | 1 | 8.420 | 6.801 | 0.95 | 0.89 | Pro only | M C L | | MA0032 | 63.16 | 587 | 594 | 653 | 660 | 1 | 7.209 | 6.986 | 0.99 | 0.97 | FREAC-3 | M C L | | M00148 | 64.86 | 589 | 595 | 655 | 661 | 1 | 6.998 | 6.998 | 0.92 | 0.92 | SRY | M C L | | MA0075 | 68.57 | 590 | 594 | 656 | 660 | 1 | 4.766 | 4.766 | 0.82 | 0.82 | S8 | M C L | | MA0080 | 66.67 | 591 | 596 | 657 | 662 | 1 | 4.391 | 4.391 | 0.82 | 0.82 | SPI-1 | M C L | | M00690* | 68.42 | 597 | 604 | 663 | 670 | 2 | 6.091 | 6.473 | 0.86 | 0.87 | Pro only | M C L | | M00974* | 63.41 | 599 | 609 | 665 | 675 | 1 | 7.650 | 7.242 | 0.84 | 0.83 | Pro only | M C L | | M00419 | 61.90 | 601 | 612 | 667 | 678 | 1 | 4.742 | 5.378 | 0.80 | 0.82 | MEIS1 | M C L | | MA0109 | 65.79 | 603 | 610 | 669 | 676 | 2 | 5.123 | 5.729 | 0.83 | 0.85 | RUSH1-alfa | M C L | | M00792* | 64.10 | 604 | 612 | 670 | 678 | 1 | 6.619 | 5.856 | 0.90 | 0.86 | Pro only | M C L | | MA0100 | 63.16 | 605 | 612 | 671 | 678 | 1 | 6.317 | 6.401 | 0.84 | 0.84 | c-MYB_1 | M C L | | M00496* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.395 | 6.035 | 0.86 | 0.89 | Pro only | M C L | | M00497* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.899 | 6.836 | 0.90 | 0.94 | Pro only | M C L | | M00498* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.533 | 4.668 | 0.88 | 0.85 | Pro only | M C L | | M00499* | 57.89 | 759 | 766 | 762 | 769 | 1 | 7.211 | 6.788 | 0.95 | 0.93 | Pro only | M C L | | M00500* | 57.89 | 759 | 766 | 762 | 769 | 1 | 5.976 | 4.838 | 0.88 | 0.84 | Pro only | M C L | | MA0080 | 55.56 | 761 | 766 | 764 | 769 | 2 | 6.483 | 8.711 | 0.91 | 1.00 | SPI-1 | M C L | | MA0098 | 55.56 | 761 | 766 | 764 | 769 | 1 | 6.300 | 7.797 | 0.93 | 1.00 | c-ETS | M C L | | M00486* | 58.97 | 771 | 779 | 774 | 782 | 1 | 5.834 | 6.213 | 0.88 | 0.90 | Pro only | M C L | | M00809* | 58.14 | 774 | 786 | 777 | 789 | 2 | 7.916 | 9.284 | 0.80 | 0.83 | Pro only | M C L | | M00967* | 61.54 | 774 | 782 | 777 | 785 | 1 | 5.262 | 5.262 | 0.80 | 0.80 | Pro only | M C L | | MA0041 | 57.14 | 775 | 786 | 778 | 789 | 2 | 7.971 | 7.289 | 0.83 | 0.81 | HFH-2 | M C L | | M00624* | 59.46 | 776 | 782 | 779 | 785 | 1 | 5.560 | 5.560 | 0.86 | 0.86 | Pro only | M C L | | M00313* | 50.00 | 792 | 799 | 795 | 802 | 2 | 5.108 | 4.395 | 0.84 | 0.81 | Pro only | M L | | M00315* | 50.00 | 792 | 799 | 795 | 802 | 2 | 4.900 | 4.234 | 0.83 | 0.80 | Pro only | M L | | M00462* | 60.00 | 807 | 816 | 815 | 824 | 2 | 5.777 | 6.024 | 0.85 | 0.86 | Pro only | M C L | | M00925* | 61.54 | 807 | 815 | 815 | 823 | 2 | 7.916 | 6.056 | 0.88 | 0.82 | Pro only | M C L | | MA0038 | 57.50 | 808 | 817 | 816 | 825 | 1 | 8.434 | 7.798 | 0.88 | 0.86 | Gfi | M C L | | M00396 | 59.46 | 809 | 815 | 817 | 823 | 2 | 3.803 | 3.803 | 0.81 | 0.81 | En-1 | M C L | | MA0035 | 58.33 | 809 | 814 | 817 | 822 | 2 | 4.265 | 4.265 | 0.85 | 0.85 | GATA-1 | M C L | | MA0037 | 58.33 | 809 | 814 | 817 | 822 | 2 | 5.162 | 5.162 | 0.86 | 0.86 | GATA-3 | M C L | | M00076 | 55.00 | 810 | 819 | 818 | 827 | 2 | 4.377 | 4.377 | 0.81 | 0.81 | GATA-2 | M C L | | M00148 | 59.46 | 810 | 816 | 818 | 824 | 1 | 5.526 | 5.526 | 0.86 | 0.86 | SRY | M C L | | M00462* | 55.00 | 810 | 819 | 818 | 827 | 2 | 4.645 | 4.645 | 0.81 | 0.81 | Pro only | M C L | | M00630* | 56.41 | 810 | 818 | 818 | 826 | 2 | 5.260 | 5.260 | 0.81 | 0.81 | Pro only | M C L | | MA0036 | 60.00 | 810 | 814 | 818 | 822 | 2 | 4.000 | 4.000 | 0.85 | 0.85 | GATA-2 | M C L | | M00285 | 51.16 | 811 | 823 | 819 | 831 | 1 | 7.570 | 6.869 | 0.88 | 0.85 | TCF11 | M C L | | M00624* | 56.76 | 811 | 817 | 819 | 825 | 1 | 4.524 | 4.524 | 0.81 | 0.81 | Pro only | M C L | | MA0089 | 58.33 | 811 | 816 | 819 | 824 | 2 | 6.811 | 6.811 | 0.91 | 0.91 | TCF11-MafG | M C L | | MA0095 | 58.33 | 811 | 816 | 819 | 824 | 1 | 5.123 | 5.123 | 0.83 | 0.83 | Yin-Yang | M C L | | M00253 | 55.26 | 812 | 819 | 820 | 827 | 1 | 6.400 | 6.400 | 0.96 | 0.96 | cap | M C L | | M00655* | 56.76 | 812 | 818 | 820 | 826 | 1 | 7.813 | 7.813 | 0.91 | 0.91 | Pro only | M C L | | M00971* | 55.26 | 812 | 819 | 820 | 827 | 1 | 5.834 | 5.834 | 0.82 | 0.82 | Pro only | M C L | | MA0035 | 58.33 | 812 | 817 | 820 | 825 | 2 | 5.498 | 5.498 | 0.91 | 0.91 | GATA-1 | M C L | | MA0036 | 60.00 | 813 | 817 | 821 | 825 | 2 | 5.978 | 5.978 | 0.96 | 0.96 | GATA-2 | M C L | | MA0098 | 58.33 | 813 | 818 | 821 | 826 | 1 | 7.229 | 7.229 | 0.97 | 0.97 | c-ETS | M C L | | M00743* | 60.53 | 821 | 828 | 829 | 836 | 1 | 6.503 | 7.812 | 0.82 | 0.87 | Pro only | M C L | | M00498* | 52.63 | 918 | 925 | 897 | 904 | 2 | 5.233 | 4.369 | 0.87 | 0.83 | Pro only | M C | | M00500* | 52.63 | 918 | 925 | 897 | 904 | 2 | 7.969 | 4.413 | 0.95 | 0.82 | Pro only | M C | | MA0080 | 50.00 | 918 | 923 | 897 | 902 | 1 | 8.579 | 4.897 | 0.99 | 0.84 | SPI-1 | M C | | M00491* | 60.47 | 922 | 934 | 901 | 913 | 2 | 8.041 | 11.563 | 0.80 | 0.88 | Pro only | M C L | | M00986* | 55.56 | 923 | 928 | 902 | 907 | 2 | 6.401 | 3.834 | 0.93 | 0.81 | Pro only | M C L | | MA0079 | 60.00 | 924 | 933 | 903 | 912 | 2 | 6.570 | 8.598 | 0.82 | 0.89 | SP1 | M C L | | M00720* | 61.54 | 925 | 933 | 904 | 912 | 2 | 7.530 | 8.270 | 0.84 | 0.87 | Pro only | M C L | | M00986* | 61.11 | 928 | 933 | 907 | 912 | 2 | 5.289 | 4.722 | 0.88 | 0.85 | Pro only | M C L | | M00428* | 63.16 | 933 | 940 | 912 | 919 | 1 | 5.146 | 4.685 | 0.85 | 0.84 | Pro only | M C L | | M00431* | 63.16 | 933 | 940 | 912 | 919 | 1 | 6.288 | 4.994 | 0.85 | 0.81 | Pro only | M C L | | M00695* | 56.76 | 937 | 943 | 916 | 922 | 2 | 7.007 | 5.823 | 0.89 | 0.84 | Pro only | M C L | | M00428* | 50.00 | 942 | 949 | 921 | 928 | 2 | 4.736 | 6.218 | 0.84 | 0.90 | Pro only | M C L | | M00803* | 50.00 | 945 | 950 | 924 | 929 | 2 | 4.808 | 4.808 | 0.81 | 0.81 | Pro only | M C L | | M00253 | 65.79 | 959 | 966 | 942 | 949 | 2 | 3.745 | 4.104 | 0.85 | 0.86 | cap | M C L | | M00468* | 64.86 | 959 | 965 | 942 | 948 | 2 | 7.266 | 6.440 | 0.94 | 0.89 | Pro only | M C L | | M00253 | 68.42 | 960 | 967 | 943 | 950 | 1 | 3.122 | 3.609 | 0.82 | 0.84 | cap | M C L | | M00313* | 68.42 | 961 | 968 | 944 | 951 | 2 | 5.673 | 5.673 | 0.87 | 0.87 | Pro only | M C L | | M00314* | 68.42 | 961 | 968 | 944 | 951 | 2 | 5.867 | 5.867 | 0.87 | 0.87 | Pro only | M C L | | M00315* | 68.42 | 961 | 968 | 944 | 951 | 2 | 5.596 | 5.596 | 0.87 | 0.87 | Pro only | M C L | | M00469* | 69.23 | 961 | 969 | 944 | 952 | 2 | 6.341 | 6.341 | 0.87 | 0.87 | Pro only | M C L | | M00470* | 69.23 | 961 | 969 | 944 | 952 | 2 | 6.454 | 6.454 | 0.85 | 0.85 | Pro only | M C L | | MA0003 | 69.23 | 961 | 969 | 944 | 952 | 2 | 6.341 | 6.341 | 0.87 | 0.87 | AP2alpha | M C L | | M00634* | 73.81 | 962 | 973 | 945 | 956 | 2 | 7.057 | 7.057 | 0.80 | 0.80 | Pro only | M C L | | MA0002 | 71.79 | 962 | 970 | 945 | 953 | 1 | 6.844 | 6.844 | 0.84 | 0.84 | AML-1 | M C L | | M00277 | 73.81 | 963 | 974 | 946 | 957 | 2 | 6.885 | 6.885 | 0.81 | 0.81 | Lmo2 complex | M C L | | MA0048 | 73.81 | 963 | 974 | 946 | 957 | 2 | 8.917 | 8.917 | 0.82 | 0.82 | Hen-1 | M C L | | M00271 | 75.00 | 964 | 969 | 947 | 952 | 1 | 5.416 | 5.416 | 0.83 | 0.83 | AML-1a | M C L | | M00253 | 76.32 | 965 | 972 | 948 | 955 | 2 | 5.465 | 5.465 | 0.92 | 0.92 | cap | M C L | | M00644* | 75.68 | 965 | 971 | 948 | 954 | 2 | 6.451 | 6.451 | 0.85 | 0.85 | Pro only | M C L | | M00695* | 75.68 | 965 | 971 | 948 | 954 | 1 | 6.047 | 6.047 | 0.85 | 0.85 | Pro only | M C L | | M00644* | 75.68 | 966 | 972 | 949 | 955 | 1 | 8.574 | 8.574 | 0.94 | 0.94 | Pro only | M C L | | M00803* | 75.00 | 970 | 975 | 953 | 958 | 2 | 4.932 | 6.310 | 0.82 | 0.88 | Pro only | M C L | | M00915* | 72.09 | 976 | 988 | 959 | 971 | 2 | 9.784 | 9.050 | 0.88 | 0.86 | Pro only | M C L | | M00322* | 75.00 | 977 | 986 | 960 | 969 | 1 | 6.101 | 7.399 | 0.81 | 0.85 | Pro only | M C L | | M00469* | 74.36 | 977 | 985 | 960 | 968 | 1 | 8.284 | 8.284 | 0.93 | 0.93 | Pro only | M C L | | M00470* | 74.36 | 977 | 985 | 960 | 968 | 1 | 8.151 | 7.624 | 0.90 | 0.89 | Pro only | M C L | | MA0003 | 74.36 | 977 | 985 | 960 | 968 | 1 | 8.284 | 8.284 | 0.93 | 0.93 | AP2alpha | M C L | | M00428* | 71.05 | 980 | 987 | 963 | 970 | 1 | 4.727 | 4.130 | 0.84 | 0.81 | Pro only | M C L | | M00803* | 72.22 | 981 | 986 | 964 | 969 | 1 | 8.895 | 8.895 | 1.00 | 1.00 | Pro only | M C L | | M00803* | 72.22 | 981 | 986 | 964 | 969 | 2 | 5.517 | 5.517 | 0.85 | 0.85 | Pro only | M C L | | M00976* | 69.23 | 981 | 989 | 964 | 972 | 1 | 5.614 | 5.961 | 0.80 | 0.82 | Pro only | M C L | | M00716* | 68.42 | 982 | 989 | 965 | 972 | 1 | 6.700 | 10.047 | 0.86 | 1.00 | Pro only | M C L | | M00803* | 66.67 | 983 | 988 | 966 | 971 | 1 | 7.517 | 7.517 | 0.94 | 0.94 | Pro only | M C L | | M00803* | 66.67 | 983 | 988 | 966 | 971 | 2 | 7.517 | 7.517 | 0.94 | 0.94 | Pro only | M C L | | M00008 | 62.50 | 990 | 999 | 976 | 985 | 1 | 9.109 | 11.207 | 0.90 | 0.97 | Sp1 | M L | | M00649* | 63.16 | 990 | 997 | 976 | 983 | 1 | 10.557 | 10.557 | 0.94 | 0.94 | Pro only | M L | | M00695* | 64.86 | 990 | 996 | 976 | 982 | 1 | 9.502 | 9.502 | 0.99 | 0.99 | Pro only | M L | | M00803* | 63.89 | 990 | 995 | 976 | 981 | 1 | 4.808 | 4.808 | 0.81 | 0.81 | Pro only | M L | | M00931* | 62.50 | 990 | 999 | 976 | 985 | 1 | 11.469 | 13.658 | 0.91 | 0.96 | Pro only | M L | | M00938* | 60.87 | 990 | 1005 | 976 | 991 | 1 | 9.502 | 9.002 | 0.83 | 0.82 | Pro only | M L | | M00986* | 63.89 | 990 | 995 | 976 | 981 | 1 | 4.722 | 4.722 | 0.85 | 0.85 | Pro only | M L | | MA0079 | 62.50 | 990 | 999 | 976 | 985 | 1 | 8.561 | 10.535 | 0.89 | 0.95 | SP1 | M L | | M00716* | 65.79 | 991 | 998 | 977 | 984 | 1 | 6.966 | 7.102 | 0.87 | 0.88 | Pro only | M L | | M00803* | 66.67 | 992 | 997 | 978 | 983 | 1 | 8.243 | 8.243 | 0.97 | 0.97 | Pro only | M L | | M00428* | 65.79 | 993 | 1000 | 979 | 986 | 2 | 7.680 | 3.958 | 0.95 | 0.81 | Pro only | M L | | M00333* | 65.12 | 994 | 1006 | 980 | 992 | 1 | 7.391 | 8.555 | 0.82 | 0.85 | Pro only | M L | | M00986* | 66.67 | 994 | 999 | 980 | 985 | 1 | 7.115 | 7.524 | 0.96 | 0.98 | Pro only | M L | | M00938* | 65.22 | 996 | 1011 | 982 | 997 | 1 | 9.979 | 8.685 | 0.84 | 0.81 | Pro only | M L | | M00716* | 68.42 | 997 | 1004 | 983 | 990 | 1 | 6.514 | 8.071 | 0.85 | 0.92 | Pro only | M L | | MA0028 | 70.00 | 998 | 1007 | 984 | 993 | 1 | 7.006 | 7.450 | 0.85 | 0.86 | Elk-1 | M L | | M00108 | 72.50 | 1000 | 1009 | 986 | 995 | 1 | 9.143 | 8.334 | 0.85 | 0.82 | NRF-2 | M L | | M00428* | 73.68 | 1000 | 1007 | 986 | 993 | 2 | 4.184 | 4.184 | 0.82 | 0.82 | Pro only | M L | | M00658* | 73.68 | 1000 | 1007 | 986 | 993 | 1 | 6.688 | 6.688 | 0.83 | 0.83 | Pro only | M L | | MA0062 | 72.50 | 1000 | 1009 | 986 | 995 | 1 | 9.143 | 8.334 | 0.85 | 0.82 | NRF-2 | M L | | MA0081 | 72.97 | 1000 | 1006 | 986 | 992 | 1 | 5.707 | 5.707 | 0.84 | 0.84 | SPI-B | M L | | M00695* | 75.68 | 1001 | 1007 | 987 | 993 | 1 | 5.454 | 5.454 | 0.83 | 0.83 | Pro only | M L | | M00971* | 73.68 | 1001 | 1008 | 987 | 994 | 2 | 8.080 | 8.080 | 0.89 | 0.89 | Pro only | M L | | M00492* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 8.448 | 7.226 | 0.91 | 0.87 | Pro only | M L | | M00496* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 5.438 | 5.517 | 0.86 | 0.87 | Pro only | M L | | M00497* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 6.977 | 7.065 | 0.95 | 0.95 | Pro only | M L | | M00498* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 4.668 | 5.233 | 0.85 | 0.87 | Pro only | M L | | M00499* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 5.727 | 5.685 | 0.89 | 0.89 | Pro only | M L | | M00500* | 73.68 | 1002 | 1009 | 988 | 995 | 2 | 5.902 | 6.673 | 0.87 | 0.90 | Pro only | M L | | M00655* | 72.97 | 1002 | 1008 | 988 | 994 | 2 | 6.124 | 6.124 | 0.84 | 0.84 | Pro only | M L | | MA0080 | 75.00 | 1002 | 1007 | 988 | 993 | 1 | 8.711 | 8.711 | 1.00 | 1.00 | SPI-1 | M L | | MA0098 | 75.00 | 1002 | 1007 | 988 | 993 | 2 | 7.797 | 7.797 | 1.00 | 1.00 | c-ETS | M L | | M00704* | 75.00 | 1003 | 1008 | 989 | 994 | 1 | 5.117 | 5.117 | 0.82 | 0.82 | Pro only | M L | | M00807* | 65.85 | 1004 | 1014 | 990 | 1000 | 1 | 7.075 | 8.516 | 0.82 | 0.86 | Pro only | M only | | MA0103 | 75.00 | 1005 | 1010 | 991 | 996 | 2 | 4.459 | 5.629 | 0.81 | 0.86 | deltaEF1 | M L | | M00695* | 67.57 | 1007 | 1013 | 993 | 999 | 1 | 5.779 | 5.663 | 0.84 | 0.84 | Pro only | M L | | M00695* | 51.35 | 1024 | 1030 | 1008 | 1014 | 1 | 9.770 | 5.779 | 1.00 | 0.84 | Pro only | M only | | M00469* | 53.85 | 1109 | 1117 | 1066 | 1074 | 2 | 7.016 | 8.905 | 0.89 | 0.95 | Pro only | M L | | M00470* | 53.85 | 1109 | 1117 | 1066 | 1074 | 2 | 7.718 | 8.735 | 0.89 | 0.92 | Pro only | M L | | MA0003 | 53.85 | 1109 | 1117 | 1066 | 1074 | 2 | 7.016 | 8.905 | 0.89 | 0.95 | AP2alpha | M L | | M00915* | 65.12 | 1131 | 1143 | 1088 | 1100 | 1 | 9.475 | 6.894 | 0.87 | 0.80 | Pro only | M C L | | M00428* | 63.16 | 1134 | 1141 | 1091 | 1098 | 2 | 5.065 | 6.548 | 0.85 | 0.91 | Pro only | M C L | | M00469* | 61.54 | 1134 | 1142 | 1091 | 1099 | 1 | 10.105 | 4.313 | 0.99 | 0.80 | Pro only | M C L | | M00469* | 61.54 | 1134 | 1142 | 1091 | 1099 | 2 | 9.691 | 8.192 | 0.98 | 0.93 | Pro only | M C L | | M00470* | 61.54 | 1134 | 1142 | 1091 | 1099 | 1 | 10.915 | 5.291 | 0.99 | 0.81 | Pro only | M C L | | M00470* | 61.54 | 1134 | 1142 | 1091 | 1099 | 2 | 10.419 | 7.835 | 0.97 | 0.89 | Pro only | M C L | | MA0003 | 61.54 | 1134 | 1142 | 1091 | 1099 | 1 | 10.105 | 4.313 | 0.99 | 0.80 | AP2alpha | M C L | | MA0003 | 61.54 | 1134 | 1142 | 1091 | 1099 | 2 | 9.691 | 8.192 | 0.98 | 0.93 | AP2alpha | M C L |
BLASTZ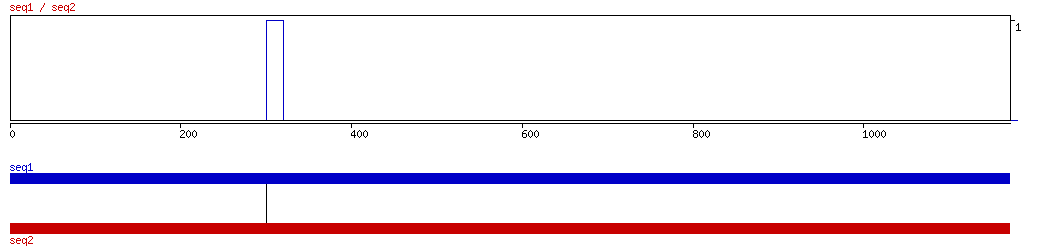
BLASTZ ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 cttgagcaaattggtggatggtggtgtcatttactgaaacggaaaatacaggatgagcagattaaaggtgaacatgcaatgaatgaaattttcaattttg
| | || | | | || | | | | | | || ||||||| ||| |
seq2 tactgaacctcttataaccttgaccatcttgtcatttaaaaaagagggtgtcaccctggtagccagtcttaaagcaaaatgaatcctcattttggaagtc
10 20 30 40 50 60 70 80 90 100
110 120 130 140 150 160 170 180 190 200
seq1 tgtatgttggtctgagatgccttttggatacatgttgtatacatgcttacatgagtaagcaactggtatgaattgacggagagggaggaaggacagggca
|| | | | | || || || | | || ||| | | | | | | | | |
seq2 tgcactcaaggaaaaaaaaaagcatgactaaatatccaacttcctagctcagtagtcgtctttcctgagtagaaaaggctcggagtgttttaatcccctt
110 120 130 140 150 160 170 180 190 200
210 220 230 240 250 260 270 280 290 300
seq1 gaagatagagctgtgggagtcatcggttccttgacagaatttacagcgtggtaatggatagactctatgctatccatctatggaacaacacatctggagA
| | | | | || || | | | | | ||| | | | | | || || ||||| | | |
seq2 ttccacgtggaagggctggggatgtcacccactgccagagtgcttgtctagtatgccacaaaggatgcttgagatgtcccggggacaacgcggtgaaacA
210 220 230 240 250 260 270 280 290 300
310 320 330 340 350 360 370 380 390 400
seq1 AGAGAGaagagctggggaggctggactgtagccttgggagtggctattaaaataggagaaggggtggagcggaagggagagaggaactatggagtgctta
| |||| || | | || | | | || ||| | | | | | | | || | | | | | |
seq2 ACTGAGaggaaaggaagggggcaagcaaaaacagaggagtaccctacaatagatgacggctagactagcccagggaaagtgcatagagaggaggaggagt
310 320 330 340 350 360 370 380 390 400
410 420 430 440 450 460 470 480 490 500
seq1 gcaagagaagaaagtgttttaaggatggggatgataaactgcattgaaagccactgagaggtggagaaaaaatcagggcagagaagtaataaaaggcatc
| | | || | || | || | | || | || ||| || | | | ||
seq2 ctgtgcacgtagttttgggacagaagggatgaaaggaagaaaaaaggtgagcaactgctctgaggataactgaaaggttgagaaaagattgacgcacaga
410 420 430 440 450 460 470 480 490 500
510 520 530 540 550 560 570 580 590 600
seq1 agcaacatggaagtcactggtgactcgcgaaaggcgatcacctttggtgtgtagggcacaaaagcctgggggggtaaagagagagtggaaagtaagaatg
| || | || | | | | | || | | | || | || ||| | | || || | | | | | |||
seq2 ataaagagggttggctcagtgggagagccttggtatgcaagaaaccgagttccacccccacaagggggcagaggaggggaaacatggctttggcaacatg
510 520 530 540 550 560 570 580 590 600
610 620 630 640 650 660 670 680 690 700
seq1 tcgagacagtcgactattattgccattttatgaatgaagaaaccgaggctcaatatttgactgtctcattcaaggtgaaagccagtggggtcagatcctg
|||| | | | | | | | | || | | | | | | | | | | |
seq2 gagagatgaagggcacagaaatttgacggggtcagtcaaagatctagagtgggcaaagtaagggtttggagacaggtgatcatcaccgacaaaccccaat
610 620 630 640 650 660 670 680 690 700
710 720 730 740 750 760 770 780 790 800
seq1 gacctagaatccatgcaatgatttgtgcacatcgcctattgaagagagagatccaattctcttccaaaacgtaaaagccaacacttccctcaaaggaggc
| | || | | | || ||| || || | |
seq2 acggagtaccaaagcagtgttgttatagggaggcgcagccacccaggagattcccgcagcgctcttccgaccttcgaaagccaaccaacccttcaactaa
710 720 730 740 750 760 770 780 790 800
810 820 830 840 850 860 870 880 890 900
seq1 ttcgagttaaatcatcctttcacgatgccgcttccattttaaagaaaaaaaaaaaaaaacgagaaggcctggtgcgggataggaatgagccgaggagcta
| | | ||| | || | | | | || ||| || | | |||| || |
seq2 aggatgcaggggcctgaaatcatccttccaagaagcagtcataaccttggagaaggccggcttcccgccctgtctgcagagcgggcgagcgcgctcgcga
810 820 830 840 850 860 870 880 890 900
910 920 930 940 950 960 970 980 990 1000
seq1 ctggccctctcggtcgggggaagacccgccccttttccgcagcccgagcccggcgcagtccactgcggctgcgcgagcctggcgcgcgcggggcgggaac
| ||| | | | | | | || | | || | ||| | | | || | | | | |
seq2 aggccccgcccctcggcgccgcgcgagcctcccctgagcgccgcactgcggctgcgccggccgggcgcgcggaggggggcggggacgcggaaggcgcggc
910 920 930 940 950 960 970 980 990 1000
1010 1020 1030 1040 1050 1060 1070 1080 1090 1100
seq1 gcggaaggggctggggttgcggcgcggcggcgaggaccagaccgggggcggggccggtagtgggagtgcggggcgcgcggtgacagcgcggggttggcgg
| || | | | | | | | | ||| | | ||| || | | | |||| || | | | || | |
seq2 ggccggggaggctgcggcggcggcggcgcggtgaagcgagagccgacgcgcggggcgaggggacgcccgacggcgtccgagggcgcggtggcgcgaggcc
1010 1020 1030 1040 1050 1060 1070 1080 1090 1100
1110 1120 1130 1140 1150 1160 1170
seq1 cgtgggacccagggggcgacagaggcagcagcagcccgaggcctgaggagaggagaccggcggcggcggca
| |||| | |
seq2 tgagggaggggacgcg-------------------------------------------------------
1110
BLASTZ ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00747* | 51.35 | 300 | 306 | 300 | 306 | 2 | 5.658 | 6.075 | 0.81 | 0.82 | Pro only | B L |
Run time: 86.8 sec | 




