GENOMIC
Mapping
3q24. View the map and BAC contig (data from UCSC genome browser).
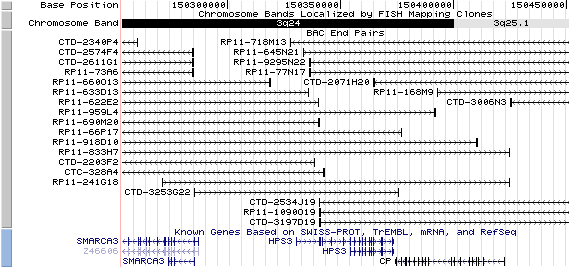
Structure
(assembly 07/03)
HPS3 (NM_032383): 17 exons, 43,936 bp, chr3:150,168,279-150,212,214.
The figure shows the structure of HPS3 gene. (data from UCSC genome browser).

Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=human HPS3, seq2=mouse Hps3) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here). Conserved regions in the upstream sequences of HPS3, HPS5 and HPS6 were identified (Stanescu, et al).
TRANSCRIPT
RefSeq/ORF
HPS3/NM_032383, 4,451 bp, view ORF and the alignment to genomic.
Note that alternate splice variants exist, but their full length sequence has not been determined.
Expression Pattern
Tissue specificity: ubiquitous. Northern blot analysis demonstrated a 4.4-kb message in all tissues tested, including heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas. Expression was greatest in kidney, with strong bands in liver and placenta.
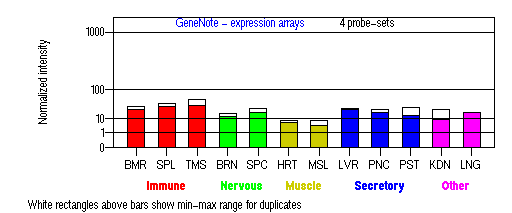
BMR: Bone marrow; SPL: Spleen; TMS: Thymus; BRN: Brain; SPC: Spinal cord; HRT: Heart; MSL: Skeletal muscle; LVR; Liver; PNC: Pancreas; PST: Prostate; KDN: Kidney; LNG: Lung. (data from GeneCards )
PROTEIN
Sequence
HPS3p (NP_115759): 1,004aa, ExPaSy NiceProt view of Swiss-Prot: Q969F9. Synonym: Hermansky-Pudlak syndrome 3 protein.
Ortholog
| Species | Mouse | Rat | Zebrafish | Drosophila |
| GeneView | coa/Hps3 | LOC310288 | ENSDARG00000015749 | CG14562 |
| Protein | NP_542365 (1,002aa) | XP_227003 (721aa) | ENSDARP00000018977 (1,001aa) | Q8MZ59 (1,190aa) |
| Identities | 79%/801aa | 64%/532aa | 47%/475aa | 17%/44aa |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc.
Domain
(1) Domains predicted by SMART:
a) ZnF NFX: 555-565
b) low complexity: 614-626
(2) Transmembrane domains predicted by SOSUI: none (predicted as a soluble protein).
Motif/Site
(1) Predicted results by ScanProsite:
a) N-glycosylation site [pattern] [Warning: pattern with a high probability of occurrence]
332-335 NLSQ, 611-614 NHSL, 854-857 NYTE, 931-934 NRTL.
b) Tyrosine sulfation site [rule] [Warning: rule with a high probability of occurrence]:
68 - 82 ayseagdYlvaieek, 402 - 416 aareedpYmdttlka, 608 - 622 fyinhslYenldeel, 848 - 862 dsladknYtedlskl.
c) cAMP- and cGMP-dependent protein kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
284 - 287 RKyS, 459 - 462 RRqS, 469 - 472 RKdT.
d) Tyrosine kinase phosphorylation site [pattern] [Warning: pattern with a high probability of occurrence]:
119 - 126 KafrDqm.Y, 296 - 304 RfapDissY.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) KDEL ER retention motif in the C-terminus: none
c) ER Membrane Retention Signals: KKXX-like motif in the C-terminus: KKPL
d) VAC possible vacuolar targeting motif: none
e) Actinin-type actin-binding motif: type 1: none; type 2: none
f) Prenylation motif: none
g) memYQRL transport motif from cell surface to Golgi: none
h) Tyrosines in the tail: too long tail
i) Dileucine motif in the tail: found LL at 994.
3D Model (updated 6/14)
(1) ModBase: none.
(2) 3D models predicted by SPARKS (fold recognition) below. View the models by PDB2MGIF.


2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=113,736, pI=6.01
FUNCTION
Ontology
a) Biological process: visual perception.
b) Plays a role in the biogenesis of lysosome-related organelles such as platelet dense granule and melanosomes (view diagram of BLOC-2 pathway here).
Location
Cytoplasmic. This gene encodes a protein containing a potential clathrin-binding motif (172-176), consensus dileucine signals, and tyrosine-based sorting signals for targeting to vesicles of lysosomal lineage.
Interaction
Gautum, et al reported that Hps3, Hps5, and Hps6 proteins regulate vesicle trafficking to lysosome-related organelles at the physiological level as components of the BLOC-2 (biogenesis of lysosome-related organelles complex-2) protein complex. The native molecular mass of BLOC-2 was estimated to be 340 +/- 64 kDa (view diagram of BLOC-2 complex here). BLOC-2 exists in a soluble pool and associates to membranes as a peripheral membrane protein (Di Pietro, et al). HPS3 associates with clathrin, predominantly on small clathrin-containing vesicles in the perinuclear region. This association most likely occurs directly via a functional clathrin-binding domain in HPS3 (Helip-Wooley, et al).
In HPS-1, lysosome associated membrane protein 1 (LAMP1), and LAMP3 were localized to abnormal large granules; in HPS-2, all LAMPs exhibited a normal granular expression; and in HPS-3, LAMP1, and LAMP3 exhibited a distinct less granular and more floccular pattern (Richmond, et al). Tyrosinase, tyrosinase-related protein-1 (Tyrp1), dopachrome tautomerase (Dct), LAMP1 and LAMP3 localization in HPS-3 melanocytes, as evaluated by immunocytochemistry and confocal microscopy, demonstrated a fine, floccular distribution in contrast to the coarse, granular distribution characteristic of control melanocytes (Boissy, et al).
No interactions found in the CuraGen database by searching its drosophila homolog CG14562.
Pathway
Involved in early stages of melanosome biogenesis and maturation (Suzuki, et al ).
MUTATION
Allele or SNP
7 mutations deposited in HGMD.
SNPs deposited in dbSNP.
7 allelic variants described in OMIM.
Distribution
| Location | Genomic | cDNA | Protein | Type | Ethnicity | Reference |
| Exon 1 | 3904bp del | Exon 1 del | N/A | gross deletion | Central Puerto Rican |
Anikster, et al |
| Exon 3 | 727insA | 727insA | I243insA | frame-shift 247X |
? | Boissy, et al |
| Intron 5 | splicing donor +1G>C |
Exon 6 del | G324_T349 del | splicing 350X |
Ashkenazi Jewish |
Huizing, et al |
| Exon 6 | 1189C>T | 1189C>T | R397W | missense | German Swiss |
Huizing, et al |
| Intron 9 | splicing donor +2T>G |
1510_1511delGT | V504delGT | splicing 504X |
Ashkenazi Jewish |
Huizing, et al |
| Intron 13 | splicing acceptor -2A>G |
Exon 14 del | I324_Q863 del | splicing | Irish German |
Huizing, et al |
| Intron 14 | splicing donor +1G>C |
Exon 14 del | I324_Q863 del | splicing | German Swiss |
Huizing, et al |
| Intron 15 | splicing acceptor -90G>A |
2798Gins 89bp | E963ins 89bp | splicing 986X |
Irish English |
Huizing, et al |
(Numbering of genomic and cDNA sequence is based on the start codon of RefSeq NM_032383.)
Effect
Anikster, et al reported that there is a central Puerto Rican founder mutation in HPS3 (3.9 kb deletion) which causes the removal of all exon 1 and 5' partial sequence of intron 1. In non-Puerto Rican patients, splicing mutation is common. The most common splicing mutation is the splicing donor mutation on intron 5 (Huizing, et al ). Most of the mutations in HPS3 are splicing mutations which may disrupt the protein function. A compound heterozygous for the 3.9 kb deletion and a I243insA mutation results in undetectable levels of the 4.4-kb HPS3 mRNA in affected cells (Boissy, et al).
PHENOTYPE
Defects in HPS3 are the cause of Hermansky-Pudlak syndrome type 3 (HPS-3, OMIM:606118). HPS-3 is an autosomal recessive disorder, characterized by oculocutaneous albinism or ocular albinism, bleeding diathesis, and lower visual acuities. Pulmonary disease is at most mild, and no patient has a history of granulomatous colitis (Huizing, et al). Patients with HPS-3 have less severe ophthalmic manifestations than patients with HPS-1 (Tsilou, et al).
REFERENCE
- Anikster Y, Huizing M, White J, Shevchenko YO, Fitzpatrick DL, Touchman JW, Compton JG, Bale SJ, Swank RT, Gahl WA, Toro JR. Mutation of a new gene causes a unique form of Hermansky-Pudlak syndrome in a genetic isolate of central Puerto Rico. Nat Genet 2001; 28: 376-80. PMID: 11455388
- Boissy RE, Richmond B, Huizing M, Helip-Wooley A, Zhao Y, Koshoffer A, Gahl WA. Melanocyte-specific proteins are aberrantly trafficked in melanocytes of Hermansky-Pudlak syndrome-type 3. Am J Pathol 2005; 166: 231-40. PMID: 15632015
- Di Pietro SM, Falcon-Perez JM, Dell'Angelica EC. Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic 2004; 5: 276-83. PMID: 15030569
- Gautam R, Chintala S, Li W, Zhang Q, Tan J, Novak EK, Di Pietro SM, Dell'Angelica EC, Swank RT. The Hermansky-Pudlak syndrome 3 (cocoa) protein is a component of the biogenesis of lysosome-related organelles complex-2 (BLOC-2). J Biol Chem 2004; 279: 12935-42. PMID: 14718540
- Helip-Wooley A, Westbroek W, Dorward H, Mommaas M, Boissy RE, Gahl WA, Huizing M. Association of the Hermansky-Pudlak syndrome type-3 protein with clathrin. BMC Cell Biol 2005; 6: 33. PMID: 16159387
- Huizing M, Anikster Y, Fitzpatrick DL, Jeong AB, D'Souza M, Rausche M, Toro JR, Kaiser-Kupfer MI, White JG, Gahl WA. Hermansky-Pudlak syndrome type 3 in Ashkenazi Jews and other non-Puerto Rican patients with hypopigmentation and platelet storage-pool deficiency. Am J Hum Genet 2001; 69: 1022-32. PMID: 11590544
- Richmond B, Huizing M, Knapp J, Koshoffer A, Zhao Y, Gahl WA, Boissy RE. Melanocytes derived from patients with Hermansky-Pudlak Syndrome types 1, 2, and 3 have distinct defects in cargo trafficking. J Invest Dermatol 2005; 124: 420-7.PMID: 15675963
- Stanescu H, Wolfsberg TG, Moreland RT, Ayub MH, Erickson E, Westbroek W, Huizing M, Gahl WA, Helip-Wooley A. Identifying putative promoter regions of Hermansky-Pudlak syndrome genes by means of phylogenetic footprinting. Ann Hum Genet 2009; 73: 422-8. PMID: 19523149
- Suzuki T, Li W, Zhang Q, Novak EK, Sviderskaya EV, Wilson A, Bennett DC, Roe BA, Swank RT, Spritz RA. The gene mutated in cocoa mice, carrying a defect of organelle biogenesis, is a homologue of the human Hermansky-Pudlak syndrome-3 gene. Genomics 2001; 78: 30-7. PMID: 11707070
- Tsilou ET, Rubin BI, Reed GF, McCain L, Huizing M, White J, Kaiser-Kupfer MI, Gahl W. Milder ocular findings in Hermansky-Pudlak syndrome type 3 compared with Hermansky-Pudlak syndrome type 1. Ophthalmology 2004; 111: 1599-603. PMID: 15288994
EDIT HISTORY:
Created by Wei Li: 06/16/2004
Updated by Wei Li: 08/18/2004
Updated by Wei Li: 11/24/2005
Updated by Wei Li: 12/25/2006
Updated by Wei Li: 07/31/2012
Updated by Wei Li: 06/13/2013