CONREAL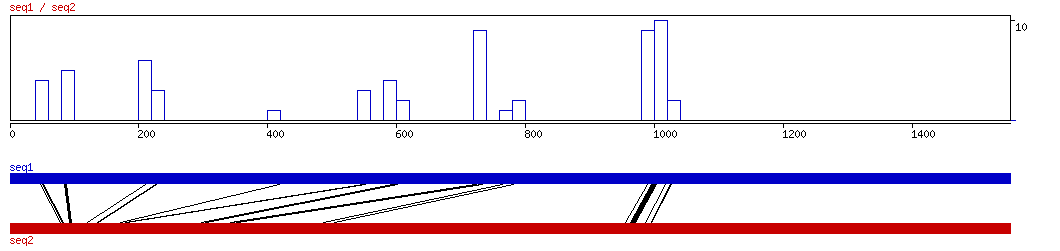
CONREAL ALIGNMENT 10 20 30 40 50 60 7
seq1 acagcatatccatttgatggaattcattcgaccactgaaagcatagt-------------------------------GAGTGAACGATCTcaacggtgg
| | || | | | | | | ||| |||| |
seq2 cccacccctcgcacccaccccccaactgagctgtgttaggcgggacagaggccacggggacagtttgctagggacaatGAGAGAACTGTGA---------
10 20 30 40 50 60 70 80 90
0 80 90 100 110 120 130 140 150 160 17
seq1 gctgagcaaagagaTGCGTGTGGCagggctatgtgctgtagatggggagttggaagaatttgccctaaggtgggctggaaggggtgggtagtgggttggg
|| |||| | | |||| | ||
seq2 --------------CCCGGGTGGTaaaagtgggtgcagggga----------------------------------------------------------
100 110
0 180 190 200 210 220 230 240 250 260 27
seq1 gggagtgatgctctctttgatctgcaggtggtggaagtatTGATTCTTagcctctgCCTTTCCCCCAagagatgctcaatctactatatccccagccagt
| |||| || || |||| | ||| | ||| | || |
seq2 ----------------------------------------TTTTTCTCagaatcgcCCTTCTCTTCAaagtttcctccttttagagcaaaa---------
120 130 140 150 160 170
0 280 290 300 310 320 330 340 350 360 37
seq1 tcttgcaacccataccttgctgggctgggctcaaagctcccaggtggcacccaggcactgaggggtggactgttcggggatttaggcccttccagagcgc
seq2 ----------------------------------------------------------------------------------------------------
0 380 390 400 410 420 430 440 450 460 47
seq1 cccctccacttccaggcacccccaatacacatacgattttttaaaataAATATTGcaaacataggttcaacacaaaactcctaatgtgcactccactccc
||| |
seq2 ------------------------------------------------TATAAAGa--------------------------------------------
0 480 490 500 510 520 530 540 550 560 57
seq1 aaaatagattgagaactcctataaattccagcgttggcactctgggggcaactgagcccttctgtgcgctatgtcacctCCCCAAAGGTcagggagagca
||||| |||| | |
seq2 -------------------------------------------------------------------------------GAACAAAGATcagaaaatgag
180 190 20
0 580 590
seq1 gtcacaaaaggacagctctagcc--------------------------------------------------------------------------CCA
|| | | ||
seq2 gttagccgcaaccgtaaaaccagatacccaccgaaaagtatgcgggtgcttaccggaagttaccatcaaggcaagcccgagaatggtgcagccaggcTCA
0 210 220 230 240 250 260 270 280 290 30
600 610 620 630 640 650 660 670 680 690
seq1 GAACTTCAGTTCAttaggcaaagcctgaggttccggggcatggggaggagagaagggatcagctgaggagccaggaaacctgggttctgagcctggttcg
|| |||| | | || | || | | |
seq2 GAGTTTCATTGCTttcaaaatccccaggatcaaaccttcctc----------------------------------------------------------
0 310 320 330 340
700 710 720 730 740 750 760
seq1 gaccctcagcaagttatttcttctctCTGCGCCTCCGTTTCCTCTtctgtaaaatggggcatgggcaa--------------------------------
|| ||||| | ||| |||| | || || |
seq2 --------------------------CTCCGCCTTAGATTCTTCTtgcctgttatcaaaatgggaggattccagaggcagcttatttctcttgttatttg
350 360 370 380 390 400 410
770 780 790
seq1 -----------------------------------------------------------------------GGCCTGGCAGgggcgggGCTCGAACccgc
| |||||| || | | || | ||
seq2 aagccgggaaataagaataacacctacccaccgttcttgtccttaagatgagcctactgggctgaggttgcGACCTGGCTGgtaggaaCCCCGTAAcccg
420 430 440 450 460 470 480 490 500 510
800 810 820 830 840 850 860 870 880 890
seq1 ggaaggcgacggcggctccggcccctcccggggcgggcggtcacgtgcgcgatcacgcggtcctacccggaagcgcgcccgggctcctgcaggcggggcg
| | | | | | | |||| || | | ||| | || | | | | | | | | |
seq2 gtaggaaggtgcaacagcgctctgttccctggcctcccaagaggagtttgaatctctgagttgaatgggaccttaggaaagcccaaccagaaccaaaccc
520 530 540 550 560 570 580 590 600 610
900 910 920 930 940 950 960 970 980
seq1 ctgtgcgcgccgcgatccggtacgtgggcctccgggctgtcccctctgggggcggcgatcctccctccggagccccccttcaaccctcccggaagt----
||| | || | ||| | || | | || | | |||| ||| ||| | | |
seq2 gaatgcagcttttcactgggacctcctgccagcttgcgatgggttgtgtagtaacccatccctgctcaccgctccctggggctgctcagggtgggattgc
620 630 640 650 660 670 680 690 700 710
seq1 ----------------------------------------------------------------------------------------------------
seq2 tgaccccacctgtgtgtgaatgagtgacccaaatataaactgtccagttgcagaggaggtaggagctaatgtgtcatcaacgcttgctccacataaagga
720 730 740 750 760 770 780 790 800 810
seq1 ----------------------------------------------------------------------------------------------------
seq2 caccttctagcttgtggtccttaaaatgggggaacctgtgacttctcagcccgtgtccaggttgtccactctcaggaacaggcggcagtggaaaccagac
820 830 840 850 860 870 880 890 900 910
990 1000 1010 1020 1030 1040
seq1 ----------------------------------------GAGGACCAGGGATGCTGTGCTGCTctcc---CATGAGccAGTCACCgagtcggtctgctg
||| ||| ||||||||||||||| | ||| | |||| | || || ||
seq2 cctctgaatgccttcatgccttttctgccttaagcatttgCAGGCCCAAGGATGCTGTGCTGCTgttgtctGCTGACctGGTCAGAgtatctagctcctc
920 930 940 950 960 970 980 990 1000 1010
1050 1060 1070 1080 1090
seq1 cagccctttctgaacctctggccgtctggatgctccactgtgcttgccaag-
| | || | | ||| | | ||
seq2 agaagccctgctgtgtgctttgccaaggctgaactcacggatgtgacctttg
1020 1030 1040 1050 1060
CONREAL ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00747* | 51.35 | 48 | 54 | 79 | 85 | 2 | 6.834 | 5.565 | 0.85 | 0.80 | Pro only | C only | | MA0080 | 52.78 | 50 | 55 | 81 | 86 | 1 | 4.376 | 4.644 | 0.82 | 0.83 | SPI-1 | C only | | M00183 | 50.00 | 50 | 59 | 81 | 90 | 1 | 5.890 | 5.769 | 0.82 | 0.82 | c-Myb | C only | | M00913* | 53.85 | 52 | 60 | 83 | 91 | 1 | 5.556 | 6.417 | 0.84 | 0.87 | Pro only | C only | | M00322* | 50.00 | 84 | 93 | 92 | 101 | 2 | 8.046 | 5.988 | 0.87 | 0.80 | Pro only | C only | | M00253 | 52.63 | 85 | 92 | 93 | 100 | 2 | 4.194 | 3.749 | 0.87 | 0.85 | cap | C only | | MA0006 | 52.78 | 86 | 91 | 94 | 99 | 1 | 5.752 | 4.891 | 0.84 | 0.80 | Ahr-ARNT | C only | | M00751* | 55.56 | 88 | 93 | 96 | 101 | 1 | 6.502 | 6.346 | 0.84 | 0.83 | Pro only | C only | | M00271 | 55.56 | 88 | 93 | 96 | 101 | 1 | 6.779 | 6.101 | 0.88 | 0.85 | AML-1a | C only | | M00500* | 57.89 | 210 | 217 | 120 | 127 | 1 | 4.080 | 5.383 | 0.81 | 0.86 | Pro only | C L M | | M00498* | 57.89 | 210 | 217 | 120 | 127 | 1 | 5.533 | 5.233 | 0.88 | 0.87 | Pro only | C L M | | M00499* | 57.89 | 210 | 217 | 120 | 127 | 1 | 4.898 | 6.782 | 0.85 | 0.93 | Pro only | C L M | | M00493* | 57.89 | 210 | 217 | 120 | 127 | 1 | 6.057 | 6.131 | 0.88 | 0.88 | Pro only | C L M | | M00494* | 57.89 | 210 | 217 | 120 | 127 | 1 | 5.247 | 5.439 | 0.84 | 0.84 | Pro only | C L M | | M00497* | 57.89 | 210 | 217 | 120 | 127 | 1 | 4.893 | 4.146 | 0.85 | 0.82 | Pro only | C L M | | MA0039 | 52.50 | 226 | 235 | 136 | 145 | 2 | 6.554 | 7.840 | 0.85 | 0.89 | Gklf | C L M | | MA0080 | 55.56 | 227 | 232 | 137 | 142 | 2 | 5.116 | 5.496 | 0.85 | 0.87 | SPI-1 | C L M | | MA0039 | 55.00 | 227 | 236 | 137 | 146 | 2 | 8.040 | 6.547 | 0.89 | 0.85 | Gklf | C L M | | M00101 | 51.35 | 418 | 424 | 171 | 177 | 1 | 3.238 | 3.440 | 0.83 | 0.84 | CdxA | C only | | M01022* | 50.00 | 549 | 558 | 179 | 188 | 1 | 6.134 | 8.714 | 0.80 | 0.88 | Pro only | C only | | M00486* | 51.28 | 550 | 558 | 180 | 188 | 1 | 4.392 | 5.540 | 0.82 | 0.87 | Pro only | C only | | M00805* | 50.00 | 551 | 556 | 181 | 186 | 1 | 6.066 | 8.180 | 0.84 | 0.94 | Pro only | C only | | M00253 | 50.00 | 593 | 600 | 297 | 304 | 1 | 4.190 | 5.344 | 0.87 | 0.92 | cap | C only | | M00314* | 50.00 | 599 | 606 | 303 | 310 | 1 | 6.669 | 4.927 | 0.91 | 0.83 | Pro only | C only | | M00313* | 50.00 | 599 | 606 | 303 | 310 | 1 | 7.075 | 5.049 | 0.94 | 0.84 | Pro only | C only | | M00315* | 50.00 | 599 | 606 | 303 | 310 | 1 | 6.968 | 4.654 | 0.93 | 0.82 | Pro only | C only | | M00747* | 51.35 | 600 | 606 | 304 | 310 | 1 | 10.025 | 5.657 | 0.97 | 0.81 | Pro only | C only | | M00253 | 52.63 | 601 | 608 | 305 | 312 | 1 | 5.500 | 5.342 | 0.92 | 0.92 | cap | C only | | MA0057 | 55.00 | 722 | 731 | 342 | 351 | 2 | 6.639 | 6.716 | 0.83 | 0.83 | MZF_5-13 | C only | | M00803* | 58.33 | 723 | 728 | 343 | 348 | 2 | 6.310 | 5.658 | 0.88 | 0.85 | Pro only | C only | | M00695* | 59.46 | 724 | 730 | 344 | 350 | 2 | 5.555 | 5.779 | 0.83 | 0.84 | Pro only | C only | | M00500* | 55.26 | 731 | 738 | 351 | 358 | 1 | 7.687 | 4.708 | 0.94 | 0.83 | Pro only | C only | | M00499* | 55.26 | 731 | 738 | 351 | 358 | 1 | 6.027 | 5.243 | 0.90 | 0.87 | Pro only | C only | | M00493* | 55.26 | 731 | 738 | 351 | 358 | 1 | 6.909 | 5.579 | 0.92 | 0.86 | Pro only | C only | | M00494* | 55.26 | 731 | 738 | 351 | 358 | 1 | 6.526 | 4.444 | 0.89 | 0.80 | Pro only | C only | | M00497* | 55.26 | 731 | 738 | 351 | 358 | 1 | 5.983 | 4.506 | 0.90 | 0.84 | Pro only | C only | | M00658* | 50.00 | 733 | 740 | 353 | 360 | 2 | 9.205 | 6.016 | 0.92 | 0.81 | Pro only | C only | | MA0092 | 50.00 | 764 | 773 | 487 | 496 | 1 | 8.157 | 7.018 | 0.86 | 0.83 | Thing1-E47 | C only | | M00431* | 52.63 | 781 | 788 | 504 | 511 | 2 | 5.024 | 4.985 | 0.81 | 0.81 | Pro only | C only | | M00428* | 52.63 | 781 | 788 | 504 | 511 | 2 | 4.677 | 4.491 | 0.84 | 0.83 | Pro only | C only | | M00967* | 53.85 | 989 | 997 | 956 | 964 | 1 | 5.970 | 5.592 | 0.83 | 0.81 | Pro only | C only | | M00076 | 60.00 | 996 | 1005 | 963 | 972 | 1 | 5.759 | 5.522 | 0.86 | 0.85 | GATA-2 | C L M | | M00075 | 60.00 | 996 | 1005 | 963 | 972 | 1 | 5.517 | 4.749 | 0.85 | 0.82 | GATA-1 | C L M | | M00253 | 57.89 | 996 | 1003 | 963 | 970 | 2 | 5.230 | 5.454 | 0.91 | 0.92 | cap | C L M | | MA0098 | 61.11 | 997 | 1002 | 964 | 969 | 2 | 5.373 | 7.229 | 0.89 | 0.97 | c-ETS | C L M | | MA0036 | 62.86 | 998 | 1002 | 965 | 969 | 1 | 5.978 | 5.978 | 0.96 | 0.96 | GATA-2 | C L M | | M00624* | 62.16 | 998 | 1004 | 965 | 971 | 2 | 6.759 | 6.759 | 0.90 | 0.90 | Pro only | C L M | | MA0035 | 61.11 | 998 | 1003 | 965 | 970 | 1 | 6.686 | 6.686 | 0.98 | 0.98 | GATA-1 | C L M | | M00253 | 63.16 | 999 | 1006 | 966 | 973 | 2 | 4.659 | 4.659 | 0.89 | 0.89 | cap | C L M | | M00253 | 63.16 | 1001 | 1008 | 968 | 975 | 2 | 2.723 | 2.723 | 0.81 | 0.81 | cap | C L M | | M00921* | 63.16 | 1002 | 1009 | 969 | 976 | 1 | 6.682 | 6.682 | 0.87 | 0.87 | Pro only | C L M | | M00624* | 64.86 | 1003 | 1009 | 970 | 976 | 2 | 5.295 | 5.295 | 0.85 | 0.85 | Pro only | C L M | | M00175 | 62.50 | 1003 | 1012 | 970 | 979 | 2 | 6.165 | 6.165 | 0.80 | 0.80 | AP-4 | C L M | | M00962* | 61.54 | 1003 | 1011 | 970 | 978 | 2 | 5.671 | 5.671 | 0.84 | 0.84 | Pro only | C L M | | M00751* | 66.67 | 1004 | 1009 | 971 | 976 | 1 | 6.346 | 6.346 | 0.83 | 0.83 | Pro only | C L M | | M00271 | 66.67 | 1004 | 1009 | 971 | 976 | 1 | 5.330 | 5.330 | 0.83 | 0.83 | AML-1a | C L M | | M00428* | 63.16 | 1004 | 1011 | 971 | 978 | 1 | 4.065 | 4.065 | 0.81 | 0.81 | Pro only | C L M | | M00253 | 63.16 | 1004 | 1011 | 971 | 978 | 2 | 4.010 | 4.010 | 0.86 | 0.86 | cap | C L M | | MA0089 | 55.56 | 1017 | 1022 | 987 | 992 | 1 | 5.823 | 5.553 | 0.87 | 0.86 | TCF11-MafG | C M | | M00801* | 52.78 | 1025 | 1030 | 995 | 1000 | 1 | 5.524 | 5.860 | 0.85 | 0.86 | Pro only | C M | | MA0089 | 50.00 | 1026 | 1031 | 996 | 1001 | 2 | 5.919 | 4.933 | 0.87 | 0.83 | TCF11-MafG | C M |
LAGAN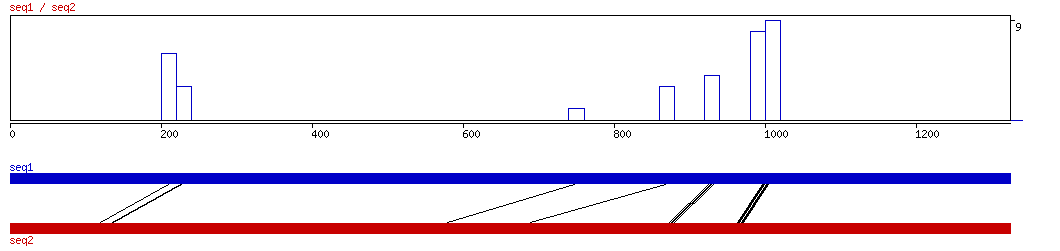
LAGAN ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 acagcatatccatttgatggaattcattcgaccactgaaagcatagtgagtgaacgatctcaacggtgggctgagcaaagagatgcgtgtggcagggcta
| | || | | || | | ||||| | || || ||| || |
seq2 cccacccctc---------gcacccaccccccaactga-------------------------------------------gctgtgttaggcgggac--
10 20 30 40
110 120 130 140 150 160 170 180 190 200
seq1 tgtgctgtagatggggagttggaagaatttgccctaaggtgggctggaaggggtgggtagtgggttggggggagtgatgctctctttgatctgcaggtgg
| || || | | ||||| ||| || || || | |||| | || ||
seq2 ----------agaggccacggggacagtttgct---agggacaatgagagaactgtgacccgggtggtaaa-----------------------agtggg
50 60 70 80 90 100 110
210 220 230 240 250 260 270 280 290 300
seq1 tggaagtatTGATTCTTagcctctgCCTTTCCCCCAagagatgctcaatctactatatccccagccagttcttgcaacccataccttgctgggctgggct
|| | | | |||| || || |||| | ||| | ||| | ||
seq2 tgcaggggaTTTTTCTCagaatcgcCCTTCTCTTCAaagtttcctccttttagag---------------------------------------------
120 130 140 150 160
310 320 330 340 350 360 370 380 390 40
seq1 caaagctcccaggtggcacccaggcactgaggggtggactgttcggggatttaggcccttccagagcgccccctccacttccaggcaccc-ccaatacac
|| | | | | | | || | | | ||| ||| | | | |||| |||| || | |
seq2 --------------------caaaatataaagagaacaaagatcagaaaatgaggtt-------agccgcaaccgtaaaaccagatacccaccgaaaagt
170 180 190 200 210 220 230 2
0 410 420 430 440 450 460
seq1 atacgattttttaaaataaat----------------------attgcaaacataggttcaaca------------caaaactcctaatgt---gcactc
|| || | ||| || | || | | ||| ||| ||||| || | | | ||
seq2 atgcgggtgcttaccggaagttaccatcaaggcaagcccgagaatggtgcagccaggctcagagtttcattgctttcaaaatccccaggatcaaaccttc
40 250 260 270 280 290 300 310 320 330 3
470 480 490 500 510 520 530 540 550 560
seq1 cactcccaaaatagattgagaactcctataaattccagcgttggcactctgggggcaactgagcccttctgtgcgctatgtcacctccccaaaggtcagg
| | || |||||| ||| | | | | | | | || | |||| || | || ||| |
seq2 ctcctccgccttagattcttcttgcctgtta-tcaaaatgggaggattccagaggcagcttatttctcttgttatttg----------------------
40 350 360 370 380 390 400 410
570 580 590 600 610 620 630 640 650 660
seq1 gagagcagtcacaaaaggacagctctagccccagaacttcagttcattaggcaaagcctgaggttccggggcatggggaggagagaagggatcagctg--
||| | | | ||| | | | | ||| ||| || ||| | ||||| | ||| ||| | ||||
seq2 --aagccgggaaataagaataacacctacccaccg--ttcttgtccttaagatgagcct-------------actgggctgaggttgcgacctggctggt
420 430 440 450 460 470 480 490 5
670 680 690 700 710 720 730 740
seq1 aggagccaggaaacctgggttctgagcctggttcggaccctcagcaagttatttcttctctctgcgcctccgtttcct----------------cttctg
|||| || | |||| || || ||| | | | ||||| || | ||| | |
seq2 aggaaccccgtaacccgg-----------------------taggaaggtgcaacagcgctctgttccctggcctcccaagaggagtttgaatctctgag
00 510 520 530 540 550 560 570
750 760 770 780 790 800 810 820 830 840
seq1 taaAATGGGgcatgggcaaggcctggcaggggcggggctcgaacccgcggaaggcgacggcggctccggcccctcccggggcgggcggtcacgtgcgcga
| |||||| | | | || ||| | | | | |||| || | || || | || ||| | || |
seq2 ttgAATGGGaccttaggaaagcccaaccagaaccaaacccgaat---------gcagcttttcactgggacctcctgccagcttgcgatgggttgtgtag
580 590 600 610 620 630 640 650 660
850 860 870 880 890 900
seq1 tcacgcggtcctacccggaagcgCGCCCGGGCtcctgcaggcggggcg-----------ctgtgcgcgc-------------------------------
| || | ||| | | || | | ||||| || || ||| ||||| | |
seq2 taacccatccctgctcacc-gctCCCTGGGGCtgctcagggtgggattgctgaccccacctgtgtgtgaatgagtgacccaaatataaactgtccagttg
670 680 690 700 710 720 730 740 750 760
910 920
seq1 --------------------------------------------------cgcgatccggtacgtgggcct-----------------------------
| | || | |||| |||
seq2 cagaggaggtaggagctaatgtgtcatcaacgcttgctccacataaaggacaccttctagcttgtggtccttaaaatgggggaacctgtgacttctcagc
770 780 790 800 810 820 830 840 850 860
930 940 950 960 970 980 990 10
seq1 ------cCGGGCTGTCCCCTCTgggg-----------gcggcgatcctccctccggagccccccttcaaccctcccggaagtg--------aggaccAGG
|| || ||||| |||| || | || | | || | || || | | | | ||| ||| |
seq2 ccgtgtcCAGGTTGTCCACTCTcaggaacaggcggcagtggaaaccagaccctctgaatgccttcatgccttttctgccttaagcatttgcaggcccAAG
870 880 890 900 910 920 930 940 950 960
00 1010 1020 1030 1040 1050 1060 1070
seq1 GATGCTGTGCTGCTctcccatg----------------agccagtcaccgagtcggtctgctgcagccctttctgaacctctggccgtctggatg-----
|||||||||||||| | || | | || | || | |||||| | || | | || | | |||||
seq2 GATGCTGTGCTGCTgttgtctgctgacctggtcagagtatctagctcctcagaagccctgctgtgtgctttgccaaggct--gaactcacggatgtgacc
970 980 990 1000 1010 1020 1030 1040 1050 1060
1080 1090
seq1 -----ctccactgtgcttgccaag
| | || | | |||
seq2 tttgccctcgcttttccctggaag
1070 1080
LAGAN ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00493* | 57.89 | 210 | 217 | 120 | 127 | 1 | 6.057 | 6.131 | 0.88 | 0.88 | Pro only | L C M | | M00494* | 57.89 | 210 | 217 | 120 | 127 | 1 | 5.247 | 5.439 | 0.84 | 0.84 | Pro only | L C M | | M00497* | 57.89 | 210 | 217 | 120 | 127 | 1 | 4.893 | 4.146 | 0.85 | 0.82 | Pro only | L C M | | M00498* | 57.89 | 210 | 217 | 120 | 127 | 1 | 5.533 | 5.233 | 0.88 | 0.87 | Pro only | L C M | | M00499* | 57.89 | 210 | 217 | 120 | 127 | 1 | 4.898 | 6.782 | 0.85 | 0.93 | Pro only | L C M | | M00500* | 57.89 | 210 | 217 | 120 | 127 | 1 | 4.080 | 5.383 | 0.81 | 0.86 | Pro only | L C M | | MA0039 | 52.50 | 226 | 235 | 136 | 145 | 2 | 6.554 | 7.840 | 0.85 | 0.89 | Gklf | L C M | | MA0039 | 55.00 | 227 | 236 | 137 | 146 | 2 | 8.040 | 6.547 | 0.89 | 0.85 | Gklf | L C M | | MA0080 | 55.56 | 227 | 232 | 137 | 142 | 2 | 5.116 | 5.496 | 0.85 | 0.87 | SPI-1 | L C M | | MA0095 | 50.00 | 748 | 753 | 579 | 584 | 2 | 5.236 | 5.236 | 0.84 | 0.84 | Yin-Yang | L M | | M00469* | 51.28 | 868 | 876 | 689 | 697 | 2 | 7.536 | 10.030 | 0.91 | 0.99 | Pro only | L only | | M00470* | 51.28 | 868 | 876 | 689 | 697 | 2 | 8.061 | 10.761 | 0.90 | 0.98 | Pro only | L only | | MA0003 | 51.28 | 868 | 876 | 689 | 697 | 2 | 7.536 | 10.030 | 0.91 | 0.99 | AP2alpha | L only | | M00253 | 50.00 | 925 | 932 | 873 | 880 | 2 | 4.480 | 2.716 | 0.88 | 0.81 | cap | L M | | M00921* | 52.63 | 928 | 935 | 876 | 883 | 1 | 5.986 | 5.826 | 0.84 | 0.84 | Pro only | L M | | M00960* | 50.00 | 929 | 938 | 877 | 886 | 2 | 9.445 | 8.655 | 0.93 | 0.90 | Pro only | L M | | M00083 | 50.00 | 932 | 939 | 880 | 887 | 2 | 9.570 | 6.396 | 0.95 | 0.82 | MZF1 | L M | | M00075 | 60.00 | 996 | 1005 | 963 | 972 | 1 | 5.517 | 4.749 | 0.85 | 0.82 | GATA-1 | L C M | | M00076 | 60.00 | 996 | 1005 | 963 | 972 | 1 | 5.759 | 5.522 | 0.86 | 0.85 | GATA-2 | L C M | | M00253 | 57.89 | 996 | 1003 | 963 | 970 | 2 | 5.230 | 5.454 | 0.91 | 0.92 | cap | L C M | | MA0098 | 61.11 | 997 | 1002 | 964 | 969 | 2 | 5.373 | 7.229 | 0.89 | 0.97 | c-ETS | L C M | | M00624* | 62.16 | 998 | 1004 | 965 | 971 | 2 | 6.759 | 6.759 | 0.90 | 0.90 | Pro only | L C M | | MA0035 | 61.11 | 998 | 1003 | 965 | 970 | 1 | 6.686 | 6.686 | 0.98 | 0.98 | GATA-1 | L C M | | MA0036 | 62.86 | 998 | 1002 | 965 | 969 | 1 | 5.978 | 5.978 | 0.96 | 0.96 | GATA-2 | L C M | | M00253 | 63.16 | 999 | 1006 | 966 | 973 | 2 | 4.659 | 4.659 | 0.89 | 0.89 | cap | L C M | | M00253 | 63.16 | 1001 | 1008 | 968 | 975 | 2 | 2.723 | 2.723 | 0.81 | 0.81 | cap | L C M | | M00921* | 63.16 | 1002 | 1009 | 969 | 976 | 1 | 6.682 | 6.682 | 0.87 | 0.87 | Pro only | L C M | | M00175 | 62.50 | 1003 | 1012 | 970 | 979 | 2 | 6.165 | 6.165 | 0.80 | 0.80 | AP-4 | L C M | | M00624* | 64.86 | 1003 | 1009 | 970 | 976 | 2 | 5.295 | 5.295 | 0.85 | 0.85 | Pro only | L C M | | M00962* | 61.54 | 1003 | 1011 | 970 | 978 | 2 | 5.671 | 5.671 | 0.84 | 0.84 | Pro only | L C M | | M00253 | 63.16 | 1004 | 1011 | 971 | 978 | 2 | 4.010 | 4.010 | 0.86 | 0.86 | cap | L C M | | M00271 | 66.67 | 1004 | 1009 | 971 | 976 | 1 | 5.330 | 5.330 | 0.83 | 0.83 | AML-1a | L C M | | M00428* | 63.16 | 1004 | 1011 | 971 | 978 | 1 | 4.065 | 4.065 | 0.81 | 0.81 | Pro only | L C M | | M00751* | 66.67 | 1004 | 1009 | 971 | 976 | 1 | 6.346 | 6.346 | 0.83 | 0.83 | Pro only | L C M |
MAVID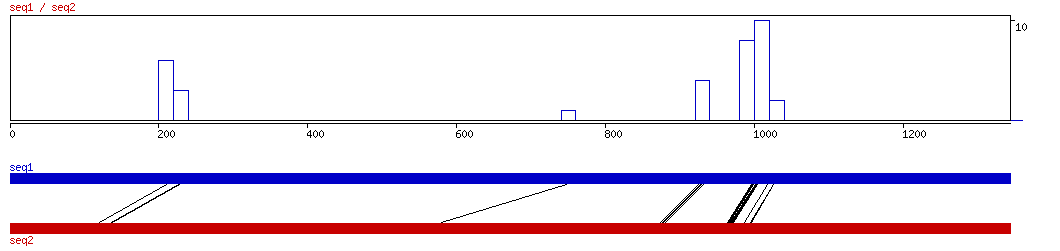
MAVID ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 acagcatatccatttgatggaattcattcgaccactgaaagcatagtgagtgaacgatctcaacggtgggctgagcaaagagatgcgtgtggcagggcta
| | || | | || | | ||||| |||| | | | | | | || || |
seq2 cccacccctc---------gcacccaccccccaactga-------------------------------gctgtgttaggcgggacagaggccacgggga
10 20 30 40 50 60
110 120 130 140 150 160 170 180 190
seq1 ---tgtgctgtagatggggagttggaagaatttgccctaaggtgggctggaaggggtgggta--gtgggttggggggagtgatgctctctttgatctgca
| |||| | || | | || | | || || |||||| | |||||| |||||
seq2 cagtttgct----agggacaatgagagaactgtgacc---------------cgggtggtaaaagtgggtgcagggga----------------------
70 80 90 100 110
200 210 220 230 240 250 260 270 280 290
seq1 ggtggtggaagtatTGATTCTTagcctctgCCTTTCCCCCAagagatgctcaatctactatatccccagccagttcttgcaacccataccttgctgggct
| |||| || || |||| | ||| | || || || |||| |||
seq2 --------------TTTTTCTCagaatcgcCCTTCTCTTCAaagtttcct-------------------ccttttagagcaaaatata----------aa
120 130 140 150 160 170
300 310 320 330 340 350 360 370
seq1 gggctcaaagctcccaggtggcacccaggcactgaggggtggactgttcggggatttaggcccttccagagcgcc-----ccctcc--------------
| | ||||| | ||| | ||||| |||| | | || | || ||| ||
seq2 gagaacaaagat-------------cagaaaatgagg------------------ttagccgcaaccgtaaaaccagatacccaccgaaaagtatgcggg
180 190 200 210 220 230 240
380 390 400 410 420 430 440 450 460
seq1 -----acttccaggcacccccaatacacatacgattttttaaaataaatattgcaaacat------aggttcaacaca---aaactcctaatgtgcac--
|| || ||| ||| || ||| ||| |||| || || |||| | ||| ||| | | ||
seq2 tgcttaccggaagttaccatcaaggcaagcccga-----------gaatggtgcagccaggctcagagtttcattgctttcaaaatccccaggatcaaac
250 260 270 280 290 300 310 320 330
470 480 490 500 510 520 530 540 550 5
seq1 --tccactcccaaaatagattgagaactcctataaattccagcgttggcactctgggggcaactgagcccttctgtgcgctatgtcacctccccaaaggt
||| | || |||||| ||| | || | | | | || | |||| || | || ||| ||| | |
seq2 cttcctcctccgccttagattcttcttgcctgt-tatcaaaatgggaggattccagaggcagcttatttctcttgt----tatttga-------------
340 350 360 370 380 390 400 410
60 570 580 590 600 610 620 630 640 65
seq1 cagggagagcagtcacaaaaggacagctc-tagccccagaacttcagttcattaggcaaagcct--------gaggttccggggcatggggaggagagaa
||| | | | ||| | | | | || || | | ||| || ||| | ||||| |||||| ||
seq2 -------agccgggaaataagaataacacctacccaccg---ttcttgtccttaagatgagcctactgggctgaggttgcg-------------------
420 430 440 450 460 470 480
0 660 670 680 690 700 710 720 730
seq1 gggatcagctg------aggagccaggaaacctgggttctgagcctggttcggaccctcagcaagttatttcttctctctgcgcctcc--------gttt
| ||| |||| || | |||| || | ||| | |||| ||| || ||| |||||| ||||
seq2 ------acctggctggtaggaaccccgtaacccgg--------------taggaaggt--gcaacagcgctctgttccctg-gcctcccaagaggagttt
490 500 510 520 530 540 550 560
740 750 760 770 780 790 800 810 820
seq1 cctcttctg--taaAATGGGgcatgggcaaggcctggcaggggcggggctcgaacccg--cggaaggcgacggcggct-----ccggcccctcccggg-g
|||| | |||||| | | | || || | |||| | | || ||| || ||| | || ||||| | |
seq2 gaatctctgagttgAATGGGaccttaggaaagc-----------------ccaaccagaaccaaacccgaatgcagcttttcactgggacctcctgccag
570 580 590 600 610 620 630 640
830 840 850 860 870 880 890 900
seq1 cgggcggtcacgtgcgcgatcacgcggtcctacccggaagcgcgccc--gggctcctgcaggcggggc-gctgtgcgcgcc-------------------
| ||| | || | | || | ||| | | | ||| ||| ||||| || || ||| |||| | | ||
seq2 cttgcgatgggttgtgtagtaacccatccctgctc---accgctccctggggctgctcagggtgggattgctgaccccacctgtgtgtgaatgagtgacc
650 660 670 680 690 700 710 720 730 740
910
seq1 ------------------------------------------------------------------------------------gcgatcc---------
| | |||
seq2 caaatataaactgtccagttgcagaggaggtaggagctaatgtgtcatcaacgcttgctccacataaaggacaccttctagcttgtggtccttaaaatgg
750 760 770 780 790 800 810 820 830 840
920 930 940 950 960 970 9
seq1 -ggtacgtgggcct-------------cCGGGCTGTCCCCTCTgggg-----gcggcgatcct-------ccctccggagccc------cccttcaaccc
|| || || | || || || ||||| |||| || ||||| | ||||| | | || || || ||
seq2 gggaacctgtgacttctcagcccgtgtcCAGGTTGTCCACTCTcaggaacaggcggcagtggaaaccagaccctctgaatgccttcatgccttttctgcc
850 860 870 880 890 900 910 920 930 940
80 990 1000 1010 1020 1030 1040 1050 1060 1070
seq1 tcccggaagtg-aggaccAGGGATGCTGTGCTGCTct---ccCATGAGccAGTCACCgagtcggtctgct--gcagccctttctgaacctctggcc-gtc
| | | || ||| ||| ||||||||||||||| | | ||| | |||| | || || || | |||||| || || | | |
seq2 ttaagcatttgcaggcccAAGGATGCTGTGCTGCTgttgtctGCTGACctGGTCAGAgtatctagctcctcagaagccctgctgtgtgctttgccaaggc
950 960 970 980 990 1000 1010 1020 1030 1040
1080 1090
seq1 tggatgctccactgtgc---ttgcc---------------aag
|| | | | |||| ||||| |||
seq2 tgaactcacggatgtgacctttgccctcgcttttccctggaag
1050 1060 1070 1080
MAVID ALIGNED HITS| Matrix | Identity | Seq1 from | Seq1 to | Seq2 from | Seq2 to | Strand | Score1 | Score2 | Rel.Score1 | Rel.Score2 | Factor | Found by:
C = CONREAL
L = LAGAN
M = MAVID
B = BLASTZ
| | M00493* | 57.89 | 210 | 217 | 120 | 127 | 1 | 6.057 | 6.131 | 0.88 | 0.88 | Pro only | M C L | | M00494* | 57.89 | 210 | 217 | 120 | 127 | 1 | 5.247 | 5.439 | 0.84 | 0.84 | Pro only | M C L | | M00497* | 57.89 | 210 | 217 | 120 | 127 | 1 | 4.893 | 4.146 | 0.85 | 0.82 | Pro only | M C L | | M00498* | 57.89 | 210 | 217 | 120 | 127 | 1 | 5.533 | 5.233 | 0.88 | 0.87 | Pro only | M C L | | M00499* | 57.89 | 210 | 217 | 120 | 127 | 1 | 4.898 | 6.782 | 0.85 | 0.93 | Pro only | M C L | | M00500* | 57.89 | 210 | 217 | 120 | 127 | 1 | 4.080 | 5.383 | 0.81 | 0.86 | Pro only | M C L | | MA0039 | 52.50 | 226 | 235 | 136 | 145 | 2 | 6.554 | 7.840 | 0.85 | 0.89 | Gklf | M C L | | MA0039 | 55.00 | 227 | 236 | 137 | 146 | 2 | 8.040 | 6.547 | 0.89 | 0.85 | Gklf | M C L | | MA0080 | 55.56 | 227 | 232 | 137 | 142 | 2 | 5.116 | 5.496 | 0.85 | 0.87 | SPI-1 | M C L | | MA0095 | 50.00 | 748 | 753 | 579 | 584 | 2 | 5.236 | 5.236 | 0.84 | 0.84 | Yin-Yang | M L | | M00253 | 50.00 | 925 | 932 | 873 | 880 | 2 | 4.480 | 2.716 | 0.88 | 0.81 | cap | M L | | M00921* | 52.63 | 928 | 935 | 876 | 883 | 1 | 5.986 | 5.826 | 0.84 | 0.84 | Pro only | M L | | M00960* | 50.00 | 929 | 938 | 877 | 886 | 2 | 9.445 | 8.655 | 0.93 | 0.90 | Pro only | M L | | M00083 | 50.00 | 932 | 939 | 880 | 887 | 2 | 9.570 | 6.396 | 0.95 | 0.82 | MZF1 | M L | | M00075 | 60.00 | 996 | 1005 | 963 | 972 | 1 | 5.517 | 4.749 | 0.85 | 0.82 | GATA-1 | M C L | | M00076 | 60.00 | 996 | 1005 | 963 | 972 | 1 | 5.759 | 5.522 | 0.86 | 0.85 | GATA-2 | M C L | | M00253 | 57.89 | 996 | 1003 | 963 | 970 | 2 | 5.230 | 5.454 | 0.91 | 0.92 | cap | M C L | | MA0098 | 61.11 | 997 | 1002 | 964 | 969 | 2 | 5.373 | 7.229 | 0.89 | 0.97 | c-ETS | M C L | | M00624* | 62.16 | 998 | 1004 | 965 | 971 | 2 | 6.759 | 6.759 | 0.90 | 0.90 | Pro only | M C L | | MA0035 | 61.11 | 998 | 1003 | 965 | 970 | 1 | 6.686 | 6.686 | 0.98 | 0.98 | GATA-1 | M C L | | MA0036 | 62.86 | 998 | 1002 | 965 | 969 | 1 | 5.978 | 5.978 | 0.96 | 0.96 | GATA-2 | M C L | | M00253 | 63.16 | 999 | 1006 | 966 | 973 | 2 | 4.659 | 4.659 | 0.89 | 0.89 | cap | M C L | | M00253 | 63.16 | 1001 | 1008 | 968 | 975 | 2 | 2.723 | 2.723 | 0.81 | 0.81 | cap | M C L | | M00921* | 63.16 | 1002 | 1009 | 969 | 976 | 1 | 6.682 | 6.682 | 0.87 | 0.87 | Pro only | M C L | | M00175 | 62.50 | 1003 | 1012 | 970 | 979 | 2 | 6.165 | 6.165 | 0.80 | 0.80 | AP-4 | M C L | | M00624* | 64.86 | 1003 | 1009 | 970 | 976 | 2 | 5.295 | 5.295 | 0.85 | 0.85 | Pro only | M C L | | M00962* | 61.54 | 1003 | 1011 | 970 | 978 | 2 | 5.671 | 5.671 | 0.84 | 0.84 | Pro only | M C L | | M00253 | 63.16 | 1004 | 1011 | 971 | 978 | 2 | 4.010 | 4.010 | 0.86 | 0.86 | cap | M C L | | M00271 | 66.67 | 1004 | 1009 | 971 | 976 | 1 | 5.330 | 5.330 | 0.83 | 0.83 | AML-1a | M C L | | M00428* | 63.16 | 1004 | 1011 | 971 | 978 | 1 | 4.065 | 4.065 | 0.81 | 0.81 | Pro only | M C L | | M00751* | 66.67 | 1004 | 1009 | 971 | 976 | 1 | 6.346 | 6.346 | 0.83 | 0.83 | Pro only | M C L | | MA0089 | 55.56 | 1017 | 1022 | 987 | 992 | 1 | 5.823 | 5.553 | 0.87 | 0.86 | TCF11-MafG | M C | | M00801* | 52.78 | 1025 | 1030 | 995 | 1000 | 1 | 5.524 | 5.860 | 0.85 | 0.86 | Pro only | M C | | MA0089 | 50.00 | 1026 | 1031 | 996 | 1001 | 2 | 5.919 | 4.933 | 0.87 | 0.83 | TCF11-MafG | M C |
BLASTZ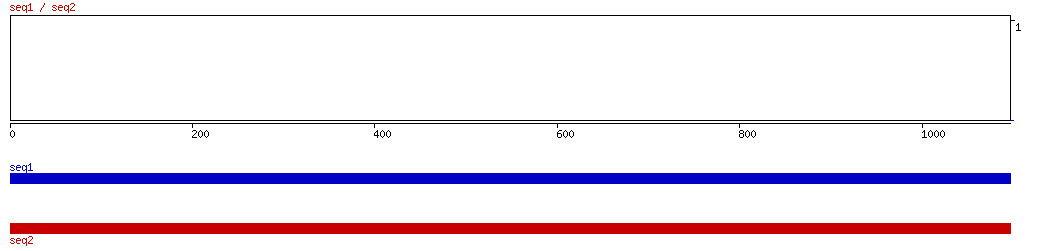
BLASTZ ALIGNMENT 10 20 30 40 50 60 70 80 90 100
seq1 acagcatatccatttgatggaattcattcgaccactgaaagcatagtgagtgaacgatctcaacggtgggctgagcaaagagatgcgtgtggcagggcta
| | || | | | | | | ||| ||| || || | | | |||
seq2 cccacccctcgcacccaccccccaactgagctgtgttaggcgggacagaggccacggggacagtttgctagggacaatgagagaactgtgacccgggtgg
10 20 30 40 50 60 70 80 90 100
110 120 130 140 150 160 170 180 190 200
seq1 tgtgctgtagatggggagttggaagaatttgccctaaggtgggctggaaggggtgggtagtgggttggggggagtgatgctctctttgatctgcaggtgg
| | || | | | | | | | | | | | ||
seq2 taaaagtgggtgcaggggatttttctcagaatcgcccttctcttcaaagtttcctccttttagagcaaaatataaagagaacaaagatcagaaaatgagg
110 120 130 140 150 160 170 180 190 200
210 220 230 240 250 260 270 280 290 300
seq1 tggaagtattgattcttagcctctgcctttcccccaagagatgctcaatctactatatccccagccagttcttgcaacccataccttgctgggctgggct
| | | | | || | | | | | | | | || | || | |
seq2 ttagccgcaaccgtaaaaccagatacccaccgaaaagtatgcgggtgcttaccggaagttaccatcaaggcaagcccgagaatggtgcagccaggctcag
210 220 230 240 250 260 270 280 290 300
310 320 330 340 350 360 370 380 390 400
seq1 caaagctcccaggtggcacccaggcactgaggggtggactgttcggggatttaggcccttccagagcgccccctccacttccaggcacccccaatacaca
| | | | || || | | | | | | | | | |
seq2 agtttcattgctttcaaaatccccaggatcaaaccttcctcctccgccttagattcttcttgcctgttatcaaaatgggaggattccagaggcagcttat
310 320 330 340 350 360 370 380 390 400
410 420 430 440 450 460 470 480 490 500
seq1 tacgattttttaaaataaatattgcaaacataggttcaacacaaaactcctaatgtgcactccactcccaaaatagattgagaactcctataaattccag
| | || || | | | | || | | || | | | | | | ||
seq2 ttctcttgttatttgaagccgggaaataagaataacacctacccaccgttcttgtccttaagatgagcctactgggctgaggttgcgacctggctggtag
410 420 430 440 450 460 470 480 490 500
510 520 530 540 550 560 570 580 590 600
seq1 cgttggcactctgggggcaactgagcccttctgtgcgctatgtcacctccccaaaggtcagggagagcagtcacaaaaggacagctctagccccagaact
| || | | | | | || ||| || || | || || |
seq2 gaaccccgtaacccggtaggaaggtgcaacagcgctctgttccctggcctcccaagaggagtttgaatctctgagttgaatgggaccttaggaaagccca
510 520 530 540 550 560 570 580 590 600
610 620 630 640 650 660 670 680 690 700
seq1 tcagttcattaggcaaagcctgaggttccggggcatggggaggagagaagggatcagctgaggagccaggaaacctgggttctgagcctggttcggaccc
| | | | || | | | || | | | | | | | | || || | || |
seq2 accagaaccaaacccgaatgcagcttttcactgggacctcctgccagcttgcgatgggttgtgtagtaacccatccctgctcaccgctccctggggctgc
610 620 630 640 650 660 670 680 690 700
710 720 730 740 750 760 770 780 790 800
seq1 tcagcaagttatttcttctctctgcgcctccgtttcctcttctgtaaaatggggcatgggcaaggcctggcaggggcggggctcgaacccgcggaaggcg
|||| | ||| || | | | | ||| | || | ||| | | | | |
seq2 tcagggtgggattgctgaccccacctgtgtgtgaatgagtgacccaaatataaactgtccagttgcagaggaggtaggagctaatgtgtcatcaacgctt
710 720 730 740 750 760 770 780 790 800
810 820 830 840 850 860 870 880 890 900
seq1 acggcggctccggcccctcccggggcgggcggtcacgtgcgcgatcacgcggtcctacccggaagcgcgcccgggctcctgcaggcggggcgctgtgcgc
| | | | | | || | |||| | | | | | | | | | || | | |||
seq2 gctccacataaaggacaccttctagcttgtggtccttaaaatgggggaacctgtgacttctcagcccgtgtccaggttgtccactctcaggaacaggcgg
810 820 830 840 850 860 870 880 890 900
910 920 930 940 950 960 970 980 990 1000
seq1 gccgcgatccggtacgtgggcctccgggctgtcccctctgggggcggcgatcctccctccggagccccccttcaaccctcccggaagtgaggaccaggga
|| | || | || | | | | | | | | || || | |
seq2 cagtggaaaccagaccctctgaatgccttcatgccttttctgccttaagcatttgcaggcccaaggatgctgtgctgctgttgtctgctgacctggtcag
910 920 930 940 950 960 970 980 990 1000
1010 1020 1030 1040 1050 1060 1070 1080 1090
seq1 tgctgtgctgctctcccatgagccagtcaccgagtcggtctgctgcagccctttctgaacctctggccgtctggatgctccactgtgcttgccaag
| ||| | | | | | | | | | | | || | | | || |
seq2 agtatctagctcctcagaagccctgctgtgtgctttgccaaggctgaactcacggatgtgacctttgccctcgcttttccctggaag---------
1010 1020 1030 1040 1050 1060 1070 1080
BLASTZ ALIGNED HITSNo hits found
Run time: 107.9 sec | 




