Results for the job "XrPsVu Wed Jul 21 17:36:00
2004":
Parameters:
PWM threshold: 80
%
Homology threshold: 50 %
Flank length: 15
bp
Plain files:
Raw
TFBSs data
List
of CONREAL aligned TFBSs
CONREAL
alignment
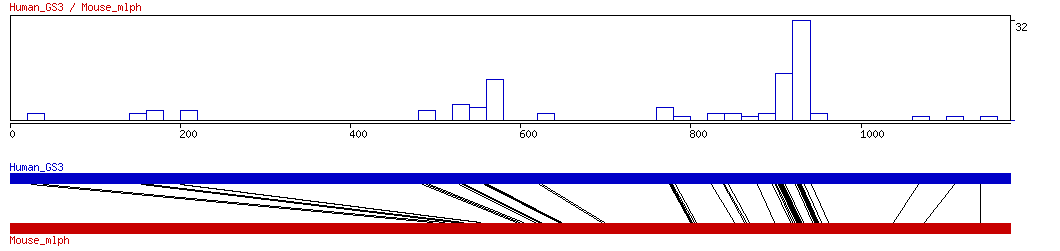
Graphical representation of aligned TFBSs.
Bars represent number of
aligned hits found within 20 bp window calculated in coordinates of the longer
sequence.
Each black line represents an aligned TFBS.
CONREAL:
CONREAL alignment:
Aligned TFBSs are shown in uppercase.
No attempts
were made to align regions not covered by TFBSs (lowercase).
10 20
Human_GS3 aagcggttcctggagaacagaatg------------------------------------
| | | | | ||
Mouse_mlphacacacacacacgcgcgcacgcgcgcgcgcgcacacacacacatctgcagatgacccaaa
10 20 30 40 50 60
Human_GS3 ------------------------------------------------------------
Mouse_mlphgcatgtcttctaaggttccctttcctctcctgctgacagggaataaatgctactatcaca
70 80 90 100 110 120
Human_GS3 ------------------------------------------------------------
Mouse_mlphtctgtccaccctggagctagttattgtttacggccctgtctctatgttgcagatgccagc
130 140 150 160 170 180
Human_GS3 ------------------------------------------------------------
Mouse_mlphactgagttgtaaaagttgtcctcaaagagtagaggcctctctctgtagctcaggctggcc
190 200 210 220 230 240
Human_GS3 ------------------------------------------------------------
Mouse_mlphtggctggaactcaatactaaaacaggttggtctgaaactttcagcaatcctcctgcttca
250 260 270 280 290 300
Human_GS3 ------------------------------------------------------------
Mouse_mlphgcctcccaagtgttgggattttgggtgtgaactagcacacacttggctaagtgaattttt
310 320 330 340 350 360
Human_GS3 ------------------------------------------------------------
Mouse_mlphgtttggttggttttttaatatagccatagggaaggggtcagggtgctcttatgctgcggc
370 380 390 400 410 420
Human_GS3 ------------------------------------------------------------
Mouse_mlphatggggggaggttatgttctttcaacatccagggcagatctctgggtggagcttctcaga
430 440 450 460 470 480
30 40 50 60 7
Human_GS3 ---------------TGAGTTTttaagtgACAAGGgagagggagggggagggagagaaaa
||||| || | ||||| ||
Mouse_mlphccacagacctactggACTGTTTtcaatagTCAAGGccttggcc-----------------
490 500 510 520
0 80 90 100 110 120 13
Human_GS3 ttctgaagcgcccagtcttggaggatgagggaggagggcaattccctaatgtccattgga
Mouse_mlph------------------------------------------------------------
0 140 150 160 170 180 19
Human_GS3 attcggaaggacaggcaggccgggcGGACTGGAGACGGCGGctcgccttgggaccctctg
||||||| | || || ||| | |
Mouse_mlph-------------------------GGACTGGGGGGGGGGGgccgcttgtgag-------
530 540 550
0 200 210 220 230 240 25
Human_GS3 agcttgcgggCCCCGCTGAGcacccaaaggccggccctagagtccaggagaagagcgcag
|| ||||| || || | || | | |
Mouse_mlph----------TCCATCTGAGggcctgggggacagcgactccagctggaaacct-------
560 570 580 590
0 260 270 280 290 300 31
Human_GS3 cggcgcggagctcccaggcgttccccgcagcgcgtcctcggtcctggaaccaccgcgccg
Mouse_mlph------------------------------------------------------------
0 320 330 340 350 360 37
Human_GS3 cgcgtcctggcttccacatctgccccatttgcccgcggatcttgactttttcttggcggg
Mouse_mlph------------------------------------------------------------
0 380 390 400 410 420 43
Human_GS3 caaggcctcggtctcctccccgcagcagggctgcccgcggctgcgggaccctcctggtcc
Mouse_mlph------------------------------------------------------------
0 440 450 460 470 480 49
Human_GS3 tgatgcctggcagaatcgaaggggtgagggtacgctctctgcagcagaaagccctGGGCT
|||||
Mouse_mlph-------------------------------------------------------GGGCT
60
0 500 510 520 530 540 55
Human_GS3 TAAAGTCCTATTtattatttggcgttgccacttactggTCGGTGGCTTTGGTAGGGCgcg
| |||| | | | | | |||| |||||| ||
Mouse_mlphCACAGTCTCACTaaaaag--------------------TTGGTGACTTTGGGTAGGGcgc
0 610 620 630 64
0 560 570 580 590 600 61
Human_GS3 tttctgagCCTCAGTTTCCCcatttgtaaagccgaagtacatagcgctttccctgcagag
||| |||||||||||| | ||| |||| | | | |
Mouse_mlphtttt----CCTCAGTTTCCCtgtctgtgaagctaagtaaagagttaaaatggatatttg-
0 650 660 670 680 690
0 620 630 640 650 660 67
Human_GS3 ttagaaatttcgACGATGTGCCGCgcgctctagagcgcgctcagcaagcgggtcccaccg
|| |||||| | ||| | | || |
Mouse_mlph------------CGGAGGTGCCGTgttctcaaaggatgcggtttgccagcaaatccctcc
700 710 720 730 740
0 680 690 700 710 720 73
Human_GS3 tgcccaaggcaggaccgcggcgcgatgtcacctatgcaaagtcgccgcaactcacgggaa
| | | | | | | | || || | |
Mouse_mlphtatgggggaccaaaacccagtggtggaaatgagcgcacaactccaacgcgtgttctg---
750 760 770 780 790
0 740 750 760 770 780 79
Human_GS3 tcgcagctgtgtgctcaactttcccctccccgccgccgtctttctCCCCAAACTGGCTTG
| | ||| ||||||
Mouse_mlph---------------------------------------------CTCTCAACCGGCTTG
800 810
0 800 810 820 830 840
Human_GS3 GGGgtcgagtatccggggctctcccccgcactgg--CGCTTCCCgcgcggGCGCCCCAGG
||| || | | | | | |||| ||| |||| | | ||| | ||
Mouse_mlphGGGacaaggtgcctctgtcctccatacccacttcagCGCCTCCCagga--GGGCCACTGG
820 830 840 850 860 870
850 860 870 880 890 900
Human_GS3 TGTCCgttccgttccgccctctgcctggcaGGGCATCCagaggtgtcc-GCGCTCCCGGA
||||| || | || | || |||| || | |||| | || |||||||
Mouse_mlphAGTCCgctctgctcttggcctgtcc-----AGGCACCCtggggtgcacgTCGATCCCGGA
880 890 900 910 920
910 920 930 940 950 960
Human_GS3 GCTGGGCGGAGCGgggCGCAGCCCACGTGGTTCGGGCGGGAGGcgccgggacgtggccag
||||||||| | || | |||||||||| | | || | | | | | |
Mouse_mlphGCTGGGCGGGTCAgg-AGTAGCCCACGTGATCTGAGCCGCAAGttgggcaggaccggcgc
930 940 950 960 970 980
970 980 990 1000 1010 1020
Human_GS3 ttgcccgcctgccccggagagccaggcgctaaccagccgctctgcgccccgcgccctgct
| | || | | | ||| ||| |
Mouse_mlphcacagctgcctggatccccgcacacagctctgtctagctcagtgcaccctg---------
990 1000 1010 1020 1030
1030 1040 1050 1060 1070 1080
Human_GS3 tgcccccattatccagccttgccccggcgccctgacctgacGCCCTGGCCTgacgccctg
| ||||||| | | ||
Mouse_mlph-----------------------------------------ACACTGGCCTcgctcggtg
1040 1050
1090 1100 1110 1120 1130
Human_GS3 cttcgtcgcctcctttctctcccAGGTGCTggaccagggactgagcgtccccc-------
| | | || || ||| ||| | | || | ||
Mouse_mlphcctggggtttctctctc------AGGAGCTaagagaaaggagcagaggccaggacccacc
1060 1070 1080 1090 1100 1110
1140 1150 1160 1170
Human_GS3 -----------------------------GGAGAGGGtccggtgtgaccccgacaagaag
|||||| | | |||||| || || || |
Mouse_mlphgagccagagctacaggctgggcattcccaGGAGAGAGgctggtgtggcctggatgaggcg
1120 1130 1140 1150 1160 1170
Human_GS3 cagaa
| ||
Mouse_mlphccgag
CONREAL aligned hits:
| Matrix |
Identity |
Seq1 from |
Seq1 to |
Seq2 from |
Seq2 to |
Strand |
Score1 |
Score2 |
Rel.Score1 |
Rel.Score2 |
Factor |
| M00148 |
0.51 |
25 |
31 |
496 |
502 |
2 |
7.141 |
4.325 |
0.92 |
0.81 |
SRY |
| M00805* |
0.53 |
39 |
44 |
510 |
515 |
1 |
5.430 |
7.031 |
0.81 |
0.88 |
Pro
only |
| M00704* |
0.50 |
155 |
160 |
524 |
529 |
1 |
5.117 |
5.117 |
0.82 |
0.82 |
Pro
only |
| M00690* |
0.50 |
157 |
164 |
526 |
533 |
2 |
6.536 |
5.276 |
0.88 |
0.83 |
Pro
only |
| M00695* |
0.54 |
161 |
167 |
530 |
536 |
1 |
5.555 |
6.198 |
0.83 |
0.86 |
Pro
only |
| M00706* |
0.56 |
162 |
170 |
531 |
539 |
1 |
7.094 |
6.736 |
0.83 |
0.82 |
Pro
only |
| M00695* |
0.59 |
164 |
170 |
533 |
539 |
1 |
6.047 |
6.198 |
0.85 |
0.86 |
Pro
only |
| M00176 |
0.53 |
200 |
209 |
552 |
561 |
2 |
9.836 |
8.070 |
0.92 |
0.85 |
AP-4 |
| M00175 |
0.53 |
200 |
209 |
552 |
561 |
2 |
9.315 |
6.870 |
0.92 |
0.83 |
AP-4 |
| M00644* |
0.51 |
201 |
207 |
553 |
559 |
2 |
5.975 |
5.975 |
0.82 |
0.82 |
Pro
only |
| M00646* |
0.50 |
485 |
492 |
595 |
602 |
1 |
6.254 |
7.363 |
0.83 |
0.87 |
Pro
only |
| M00805* |
0.56 |
489 |
494 |
599 |
604 |
1 |
5.774 |
5.774 |
0.83 |
0.83 |
Pro
only |
| M00630* |
0.59 |
493 |
501 |
603 |
611 |
2 |
6.978 |
5.260 |
0.87 |
0.81 |
Pro
only |
| M00792* |
0.56 |
528 |
536 |
618 |
626 |
2 |
5.040 |
5.804 |
0.82 |
0.85 |
Pro
only |
| M00727* |
0.58 |
532 |
539 |
622 |
629 |
1 |
7.195 |
7.940 |
0.84 |
0.87 |
Pro
only |
| M00486* |
0.56 |
533 |
541 |
623 |
631 |
2 |
7.190 |
7.293 |
0.94 |
0.94 |
Pro
only |
| M00159 |
0.51 |
534 |
546 |
624 |
636 |
1 |
6.069 |
6.958 |
0.83 |
0.86 |
C/EBP |
| M00805* |
0.56 |
535 |
540 |
625 |
630 |
2 |
6.098 |
6.098 |
0.84 |
0.84 |
Pro
only |
| M00801* |
0.64 |
558 |
563 |
644 |
649 |
1 |
4.845 |
4.845 |
0.82 |
0.82 |
Pro
only |
| M00670* |
0.66 |
559 |
566 |
645 |
652 |
1 |
6.475 |
6.475 |
0.83 |
0.83 |
Pro
only |
| M00747* |
0.65 |
559 |
565 |
645 |
651 |
1 |
6.075 |
6.075 |
0.82 |
0.82 |
Pro
only |
| M00183 |
0.62 |
559 |
568 |
645 |
654 |
2 |
5.361 |
5.361 |
0.80 |
0.80 |
c-Myb |
| M00148 |
0.65 |
560 |
566 |
646 |
652 |
2 |
6.186 |
6.186 |
0.88 |
0.88 |
SRY |
| M00415 |
0.62 |
561 |
569 |
647 |
655 |
1 |
5.770 |
5.770 |
0.83 |
0.83 |
AREB6 |
| M00486* |
0.62 |
561 |
569 |
647 |
655 |
2 |
5.562 |
5.562 |
0.87 |
0.87 |
Pro
only |
| M00492* |
0.61 |
561 |
568 |
647 |
654 |
1 |
5.424 |
5.424 |
0.80 |
0.80 |
Pro
only |
| M00497* |
0.61 |
561 |
568 |
647 |
654 |
1 |
3.745 |
3.745 |
0.80 |
0.80 |
Pro
only |
| M00492* |
0.63 |
562 |
569 |
648 |
655 |
1 |
6.087 |
6.087 |
0.83 |
0.83 |
Pro
only |
| M00500* |
0.63 |
562 |
569 |
648 |
655 |
1 |
7.384 |
7.384 |
0.93 |
0.93 |
Pro
only |
| M00499* |
0.63 |
562 |
569 |
648 |
655 |
1 |
5.050 |
5.050 |
0.86 |
0.86 |
Pro
only |
| M00493* |
0.63 |
562 |
569 |
648 |
655 |
1 |
6.385 |
6.385 |
0.89 |
0.89 |
Pro
only |
| M00496* |
0.63 |
562 |
569 |
648 |
655 |
1 |
6.972 |
6.972 |
0.93 |
0.93 |
Pro
only |
| M00494* |
0.63 |
562 |
569 |
648 |
655 |
1 |
6.159 |
6.159 |
0.88 |
0.88 |
Pro
only |
| M00497* |
0.63 |
562 |
569 |
648 |
655 |
1 |
6.345 |
6.345 |
0.92 |
0.92 |
Pro
only |
| M00750* |
0.62 |
562 |
568 |
648 |
654 |
2 |
6.728 |
6.728 |
0.88 |
0.88 |
Pro
only |
| M00655* |
0.51 |
622 |
628 |
695 |
701 |
2 |
6.158 |
6.553 |
0.84 |
0.86 |
Pro
only |
| M00428* |
0.50 |
626 |
633 |
699 |
706 |
1 |
7.661 |
4.837 |
0.95 |
0.84 |
Pro
only |
| M00004 |
0.50 |
775 |
792 |
800 |
817 |
2 |
8.001 |
8.871 |
0.81 |
0.83 |
c-Myb |
| M00690* |
0.53 |
776 |
783 |
801 |
808 |
1 |
7.538 |
6.201 |
0.91 |
0.86 |
Pro
only |
| M00183 |
0.53 |
777 |
786 |
802 |
811 |
1 |
8.781 |
8.328 |
0.94 |
0.92 |
c-Myb |
| M00773* |
0.54 |
779 |
789 |
804 |
814 |
2 |
7.479 |
6.907 |
0.89 |
0.86 |
Pro
only |
| M00072 |
0.54 |
782 |
792 |
807 |
817 |
2 |
9.845 |
6.413 |
0.91 |
0.80 |
CP2 |
| M00497* |
0.50 |
824 |
831 |
851 |
858 |
1 |
7.974 |
4.174 |
1.00 |
0.82 |
Pro
only |
| M00428* |
0.58 |
838 |
845 |
863 |
870 |
2 |
4.531 |
5.138 |
0.83 |
0.85 |
Pro
only |
| M00469* |
0.59 |
840 |
848 |
865 |
873 |
1 |
8.151 |
4.365 |
0.93 |
0.80 |
Pro
only |
| M00792* |
0.56 |
844 |
852 |
869 |
877 |
2 |
5.660 |
6.155 |
0.84 |
0.87 |
Pro
only |
| M00497* |
0.61 |
878 |
885 |
898 |
905 |
2 |
5.255 |
3.807 |
0.87 |
0.80 |
Pro
only |
| M00087 |
0.60 |
896 |
907 |
917 |
928 |
2 |
6.036 |
7.575 |
0.80 |
0.85 |
Ik-2 |
| M00800* |
0.59 |
899 |
914 |
920 |
935 |
1 |
7.275 |
7.275 |
0.81 |
0.81 |
Pro
only |
| M00002 |
0.58 |
901 |
915 |
922 |
936 |
1 |
8.360 |
8.360 |
0.81 |
0.81 |
E47 |
| M00189 |
0.60 |
903 |
914 |
924 |
935 |
2 |
6.978 |
6.978 |
0.82 |
0.82 |
AP-2 |
| M00176 |
0.57 |
903 |
912 |
924 |
933 |
1 |
7.355 |
7.355 |
0.83 |
0.83 |
AP-4 |
| M00175 |
0.57 |
903 |
912 |
924 |
933 |
1 |
7.089 |
7.089 |
0.84 |
0.84 |
AP-4 |
| M00175 |
0.57 |
903 |
912 |
924 |
933 |
2 |
6.386 |
6.386 |
0.81 |
0.81 |
AP-4 |
| M00644* |
0.62 |
904 |
910 |
925 |
931 |
2 |
6.737 |
6.737 |
0.86 |
0.86 |
Pro
only |
| M00720* |
0.59 |
904 |
912 |
925 |
933 |
1 |
8.036 |
8.036 |
0.86 |
0.86 |
Pro
only |
| M00644* |
0.59 |
905 |
911 |
926 |
932 |
1 |
5.975 |
5.975 |
0.82 |
0.82 |
Pro
only |
| M00196 |
0.56 |
907 |
919 |
928 |
940 |
1 |
12.140 |
9.614 |
0.89 |
0.84 |
Sp1 |
| M00430* |
0.63 |
908 |
915 |
929 |
936 |
1 |
6.593 |
6.593 |
0.82 |
0.82 |
Pro
only |
| M00428* |
0.63 |
908 |
915 |
929 |
936 |
1 |
3.824 |
3.824 |
0.80 |
0.80 |
Pro
only |
| M00807* |
0.59 |
908 |
918 |
929 |
939 |
1 |
8.726 |
7.902 |
0.87 |
0.84 |
Pro
only |
| M00695* |
0.65 |
909 |
915 |
930 |
936 |
1 |
7.466 |
7.466 |
0.91 |
0.91 |
Pro
only |
| M00008 |
0.60 |
909 |
918 |
930 |
939 |
1 |
7.228 |
7.401 |
0.84 |
0.84 |
Sp1 |
| M00803* |
0.64 |
911 |
916 |
932 |
937 |
1 |
5.589 |
8.271 |
0.84 |
0.97 |
Pro
only |
| M00615* |
0.50 |
923 |
942 |
943 |
962 |
1 |
12.598 |
10.780 |
0.90 |
0.85 |
Pro
only |
| M00615* |
0.50 |
923 |
942 |
943 |
962 |
2 |
12.598 |
10.780 |
0.90 |
0.85 |
Pro
only |
| M00236 |
0.54 |
925 |
940 |
945 |
960 |
1 |
8.113 |
9.834 |
0.82 |
0.85 |
Arnt |
| M00236 |
0.54 |
925 |
940 |
945 |
960 |
2 |
10.737 |
10.732 |
0.87 |
0.87 |
Arnt |
| M00624* |
0.62 |
926 |
932 |
946 |
952 |
1 |
4.879 |
4.879 |
0.83 |
0.83 |
Pro
only |
| M00118 |
0.57 |
926 |
939 |
946 |
959 |
1 |
13.393 |
10.928 |
0.89 |
0.85 |
c-Myc/Max |
| M00118 |
0.57 |
926 |
939 |
946 |
959 |
2 |
13.393 |
10.928 |
0.89 |
0.85 |
c-Myc/Max |
| M00119 |
0.57 |
926 |
939 |
946 |
959 |
1 |
12.624 |
12.369 |
0.91 |
0.90 |
Max |
| M00119 |
0.57 |
926 |
939 |
946 |
959 |
2 |
12.624 |
12.369 |
0.91 |
0.90 |
Max |
| M00121 |
0.57 |
926 |
939 |
946 |
959 |
1 |
11.954 |
13.559 |
0.91 |
0.94 |
USF |
| M00121 |
0.57 |
926 |
939 |
946 |
959 |
2 |
11.954 |
13.559 |
0.91 |
0.94 |
USF |
| M00122 |
0.57 |
926 |
939 |
946 |
959 |
1 |
9.894 |
11.105 |
0.91 |
0.94 |
USF |
| M00122 |
0.57 |
926 |
939 |
946 |
959 |
2 |
9.894 |
11.105 |
0.91 |
0.94 |
USF |
| M00470* |
0.59 |
927 |
935 |
947 |
955 |
1 |
5.561 |
5.561 |
0.82 |
0.82 |
Pro
only |
| M00469* |
0.59 |
927 |
935 |
947 |
955 |
1 |
5.200 |
5.200 |
0.83 |
0.83 |
Pro
only |
| M00123 |
0.55 |
927 |
938 |
947 |
958 |
1 |
8.987 |
8.085 |
0.86 |
0.83 |
c-Myc/Max |
| M00123 |
0.55 |
927 |
938 |
947 |
958 |
2 |
10.843 |
9.699 |
0.91 |
0.88 |
c-Myc/Max |
| M00055 |
0.55 |
927 |
938 |
947 |
958 |
1 |
10.332 |
10.546 |
0.90 |
0.91 |
N-Myc |
| M00055 |
0.55 |
927 |
938 |
947 |
958 |
2 |
10.003 |
8.111 |
0.89 |
0.84 |
N-Myc |
| M00796* |
0.55 |
927 |
938 |
947 |
958 |
1 |
9.403 |
11.550 |
0.87 |
0.92 |
Pro
only |
| M00796* |
0.55 |
927 |
938 |
947 |
958 |
2 |
12.204 |
10.216 |
0.94 |
0.89 |
Pro
only |
| M00468* |
0.59 |
928 |
934 |
948 |
954 |
2 |
5.427 |
5.427 |
0.84 |
0.84 |
Pro
only |
| M00184 |
0.55 |
928 |
937 |
948 |
957 |
1 |
6.299 |
6.299 |
0.80 |
0.80 |
MyoD |
| M00322* |
0.55 |
928 |
937 |
948 |
957 |
1 |
9.148 |
8.161 |
0.90 |
0.87 |
Pro
only |
| M00322* |
0.55 |
928 |
937 |
948 |
957 |
2 |
9.629 |
7.533 |
0.92 |
0.85 |
Pro
only |
| M00187 |
0.55 |
928 |
937 |
948 |
957 |
1 |
9.850 |
9.249 |
0.92 |
0.90 |
USF |
| M00187 |
0.55 |
928 |
937 |
948 |
957 |
2 |
8.368 |
8.865 |
0.87 |
0.88 |
USF |
| M00799* |
0.59 |
929 |
935 |
949 |
955 |
2 |
10.927 |
10.927 |
1.00 |
1.00 |
Pro
only |
| M00726* |
0.61 |
930 |
935 |
950 |
955 |
1 |
8.133 |
8.133 |
1.00 |
1.00 |
Pro
only |
| M00726* |
0.61 |
930 |
935 |
950 |
955 |
2 |
8.133 |
8.133 |
1.00 |
1.00 |
Pro
only |
| M00799* |
0.59 |
930 |
936 |
950 |
956 |
1 |
10.927 |
7.811 |
1.00 |
0.89 |
Pro
only |
| M00396 |
0.57 |
933 |
939 |
953 |
959 |
1 |
4.504 |
4.063 |
0.85 |
0.83 |
En-1 |
| M00431* |
0.50 |
942 |
949 |
962 |
969 |
2 |
6.173 |
5.815 |
0.84 |
0.83 |
Pro
only |
| M00428* |
0.50 |
942 |
949 |
962 |
969 |
2 |
5.955 |
6.332 |
0.89 |
0.90 |
Pro
only |
| M00761* |
0.53 |
1068 |
1077 |
1038 |
1047 |
2 |
5.167 |
6.054 |
0.80 |
0.83 |
Pro
only |
| M00624* |
0.51 |
1110 |
1116 |
1074 |
1080 |
2 |
5.583 |
4.314 |
0.86 |
0.81 |
Pro
only |
| M00649* |
0.71 |
1140 |
1147 |
1140 |
1147 |
1 |
8.703 |
6.920 |
0.87 |
0.81 |
Pro
only |
