GENOMIC
Mapping
15qE1. View the map and BAC clones (data from UCSC genome browser).
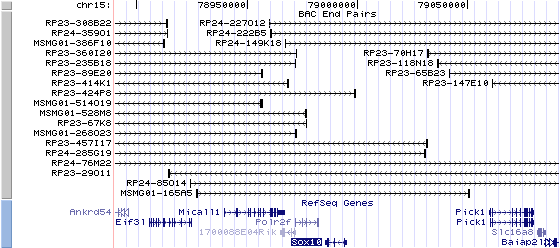
Structure
(assembly 02/2006)
Sox10 (NM_011437): 4 exons, 9,577 bp, chr15:78,985,344-78,994,920.
The figure below shows the structure of the Sox10 gene (data from UCSC genome browser).
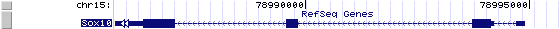
Regulatory Element
Search the 5'UTR and 1kb upstream regions (seq1=mouse Sox10, seq2=human SOX10) by CONREAL with 80% Position Weight Matrices (PWMs) threshold (view results here).
TRANSCRIPT
RefSeq/ORF
Sox10( NM_011437), 2,713 bp, view ORF and the alignment to genomic.
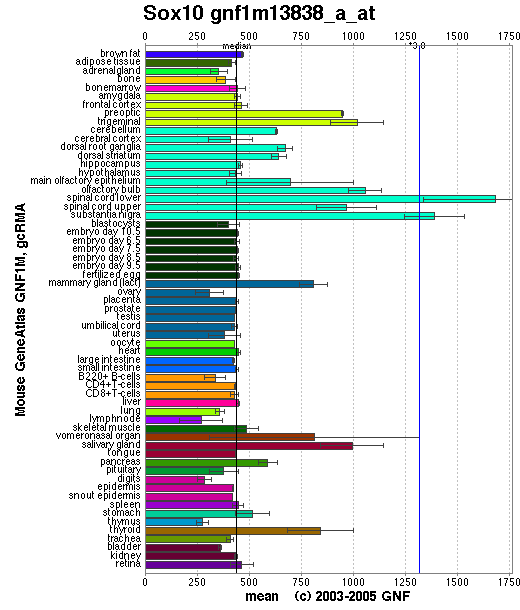
Expression Pattern
Affymetrix microarray expression pattern in SymAtlas from GNF is shown below.
Browse more information in Entrez Gene, UCSC Gene Sorter, MGI.
PROTEIN
Sequence
Transcription factor SOX-10 (SOX-21) (Transcription factor SOX-M)(NP_035567): 466 aa, UniProtKB/Swiss-Prot entry Q04888.
Ortholog
Species Human Chimpanzee Dog Rat Chicken GeneView
SOX10
SOX10
LOC481258
Sox10
SOX-10
Protein
NP_008872 (466 aa)
XP_525590 (302 aa)
XP_538379 (621 aa)
NP_062066 (466 aa)
NP_990123 (461 aa)
Identities
459/466 (98%) 278/283 (98%) 534/557 (95%) 461/466 (98%) 385/470 (81%)
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) low complexity: 4-14
b) low complexity: 32-51
c) low complexity: 87-101
d) low complexity: 126-152
e) HMG: 206-276
f) low complexity: 286-308
g) low complexity: 341-348
h) low complexity: 413-435
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 249 to 252 NESD.
b) Protein kinase C phosphorylation site:
Site : 68 to 70 SPR.
Site : 251 to 253 SDK.
Site : 347 to 349 TPK.
Site : 354 to 356 SGK.
Site : 534 to 536 SQR.
c) Casein kinase II phosphorylation site:
Site : 56 to 59 TPWD.
Site : 111 to 114 SEVE.
Site : 184 to 187 SGYD.
Site : 365 to 368 SLGE.
Site : 390 to 393 SNME.
Site : 398 to 401 TELD.
Site : 556 to 559 THWE.
d) Tyrosine kinase phosphorylation site:
Site : 269 to 276 KDHPDYKY.
e) N-myristoylation si:
Site : 101 to 106 GSDMAE.
Site : 129 to 134 GSAPSL.
Site : 138 to 143 GGGGGS.
Site : 139 to 144 GGGGSG.
Site : 140 to 145 GGGSGL.
Site : 199 to 204 GASKSK.
Site : 301 to 306 GGAAAI.
Site : 323 to 328 GSPMSD.
Site : 414 to 419 GSYSAA.
Site : 422 to 427 GLGSAL.
Site : 424 to 429 GSALAV.
Site : 519 to 524 GQASGL.
Site : 523 to 528 GLYSAF.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1)
ModBase predicted 3D structure of Q04888 from UCSC Genome Sorter:
(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=49,949Da.
| Species | Human | Chimpanzee | Dog | Rat | Chicken |
| GeneView | SOX10 | SOX10 | LOC481258 | Sox10 | SOX-10 |
| Protein | NP_008872 (466 aa) | XP_525590 (302 aa) | XP_538379 (621 aa) | NP_062066 (466 aa) | NP_990123 (461 aa) |
| Identities | 459/466 (98%) | 278/283 (98%) | 534/557 (95%) | 461/466 (98%) | 385/470 (81%) |
View multiple sequence alignment (PDF file) by ClustalW and GeneDoc. View evolutionary tree by TreeView.
Domain
(1) Domains predicted by SMART:
a) low complexity: 4-14
b) low complexity: 32-51
c) low complexity: 87-101
d) low complexity: 126-152
e) HMG: 206-276
f) low complexity: 286-308
g) low complexity: 341-348
h) low complexity: 413-435
(2) Transmembrane domains predicted by SOSUI:
This amino acid sequence is of a SOLUBLE PROTEIN.
(3) Graphic view of InterPro domain structure.
Motif/Site
(1) Predicted results by ScanProsite:
a)N-glycosylation site:
Site : 249 to 252 NESD.
b) Protein kinase C phosphorylation site:
Site : 68 to 70 SPR.
Site : 251 to 253 SDK.
Site : 347 to 349 TPK.
Site : 354 to 356 SGK.
Site : 534 to 536 SQR.
c) Casein kinase II phosphorylation site:
Site : 56 to 59 TPWD.
Site : 111 to 114 SEVE.
Site : 184 to 187 SGYD.
Site : 365 to 368 SLGE.
Site : 390 to 393 SNME.
Site : 398 to 401 TELD.
Site : 556 to 559 THWE.
d) Tyrosine kinase phosphorylation site:
Site : 269 to 276 KDHPDYKY.
e) N-myristoylation si:
Site : 101 to 106 GSDMAE.
Site : 129 to 134 GSAPSL.
Site : 138 to 143 GGGGGS.
Site : 139 to 144 GGGGSG.
Site : 140 to 145 GGGSGL.
Site : 199 to 204 GASKSK.
Site : 301 to 306 GGAAAI.
Site : 323 to 328 GSPMSD.
Site : 414 to 419 GSYSAA.
Site : 422 to 427 GLGSAL.
Site : 424 to 429 GSALAV.
Site : 519 to 524 GQASGL.
Site : 523 to 528 GLYSAF.
(2) Predicted results of subprograms by PSORT II:
a) N-terminal signal peptide: none
b) Tentative number of TMS(s) for the threshold 0.5: 0
c) KDEL ER retention motif in C-terminus: none
d) ER membrane retention signals: none
e) VAC possible vacuolar targeting motif: none
f) Actinin-type actin-binding motif: type 1: none; type 2: none
g) Prenylation motif: none
h) memYQRL transport motif from cell surface to Golgi: none
i) Tyrosines in the tail: none
j) Dileucine motif in the tail: none
3D Model
(1) ModBase predicted 3D structure of Q04888 from UCSC Genome Sorter:



(2) 3D structures are predicted by SPARKS2 and viewed by Protein Explorer.
2D-PAGE
This protein does not exist in the current release of SWISS-2DPAGE.
Computed theoretical MW=49,949Da.
FUNCTION
Ontology
(1) Biological process: DNA binding, DNA-dependent regulation of transcription, sensory perception of sound.
(2) Melanocyte differentiation.
(3) Gut morphogenesis.
(4) Enteric and peripheral nervous system development.
Location
Nucleus/Cytoplasm
Interaction
Transcription factor SOX-10. Transcription factor that seems to function synergistically with the POU domain protein TST-1/OCT6/SCIP. This gene encodes a member of the SOX (SRY-related HMG-box) family of transcription factors involved in the regulation of embryonic development and in the determination of the cell fate. The encoded protein may act as a transcriptional activator after forming a protein complex with other proteins.
Interactions in HPRD
View co-occured partners in literature searched by PPI Finder.
Pathway
SRY related HMG box gene 10,expressed in neural crest cells during early development and in glial cells of the peripheral and central nervous system during late development and in adult, playing an essential role in the development of neural-crest-derived human cell lineages, homolog to mouse dominant megacolon (Dom).
MUTATION
Allele or SNP
9 phenotypic alleles of Dom are described in MGI:98358.
SNPs deposited in dbSNP Build 128.
Distribution
The 9 phenotypic alleles include 5 targeted and other, 1 spontaneous, 1 chemically induced, and 2 radiation induced.
(Numbering of cDNA sequence is based on the start codon of RefSeq NM_011437. view ORF here.)
Effect
Mutations in this gene result in dominant megacolon.
PHENOTYPE
Defects in Sox10 are the cause of the mouse mutant dominant megacolon (dom). Homozygotes for null mutations lack peripheral glial cells, melanocytes, and autonomic and enteric neurons, and die neonatally or sooner. Heterozygotes exhibit white spotting and megacolon.
During N-ethyl-N-nitrosourea (ENU) mutagenesis screen utilizing a mouse model sensitized for NC defects, Sox10(LacZ/+), Erbb3(msp1) that carries a single amino acid substitution in the extracellular region of the protein shows altered neural crest patterning (Buac et al.). This group also identified that a null mutation of Gli3 causes the increased hypopigmentation in Sox10(LacZ/+) (Matera et al.).
REFERENCE
- Buac K, Watkins-Chow DE, Loftus SK, Larson DM, Incao A, Gibney G, Pavan WJ. A Sox10 expression screen identifies an amino acid essential for Erbb3 function. PLoS Genet 2008; 4:e1000177. PMID: 18773073
- Matera I, Watkins-Chow DE, Loftus SK, Hou L, Incao A, Silver DL, Rivas C, Elliott EC, Baxter LL, Pavan WJ. A sensitized mutagenesis screen identifies Gli3 as a modifier of Sox10 neurocristopathy. Hum Mol Genet 2008; 17:2118-31. PMID: 18397875